Fri, Apr 26, 2024
[Archive]
Volume 20, Issue 8 (August 2022)
IJRM 2022, 20(8): 671-682 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Yousefian M, Angaji A, Siasi E, Rahmani S A, Abbasalizadeh Khiaban S. Role of CYP1A1, CYP2D6, and NOS3 gene polymorphisms in idiopathic recurrent pregnancy loss in the Iranian Azeri population: A case-control study. IJRM 2022; 20 (8) :671-682
URL: http://ijrm.ir/article-1-2151-en.html
URL: http://ijrm.ir/article-1-2151-en.html
Mahsa Yousefian1 

 , Abdolhamid Angaji *
, Abdolhamid Angaji * 

 2, Elham Siasi1
2, Elham Siasi1 

 , Seyed Ali Rahmani3
, Seyed Ali Rahmani3 

 , Shamsi Abbasalizadeh Khiaban4
, Shamsi Abbasalizadeh Khiaban4 




 , Abdolhamid Angaji *
, Abdolhamid Angaji * 

 2, Elham Siasi1
2, Elham Siasi1 

 , Seyed Ali Rahmani3
, Seyed Ali Rahmani3 

 , Shamsi Abbasalizadeh Khiaban4
, Shamsi Abbasalizadeh Khiaban4 


1- Department of Genetics, Faculty of Biological Sciences, North Tehran Branch, Islamic Azad University, Tehran, Iran.
2- Department of Cell and Molecular Biology, Faculty of Biological Sciences, Kharazmi University, Tehran, Iran. , Angaji@khu.ac.ir
3- Department of Medical Genetics, School of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
4- Department of Obstetrics and Gynecology, Women’s Reproductive Health Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
2- Department of Cell and Molecular Biology, Faculty of Biological Sciences, Kharazmi University, Tehran, Iran. , Angaji@khu.ac.ir
3- Department of Medical Genetics, School of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
4- Department of Obstetrics and Gynecology, Women’s Reproductive Health Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
Full-Text [PDF 1199 kb]
(535 Downloads)
| Abstract (HTML) (730 Views)
1. Introduction
Pregnancy loss is defined as a loss of pregnancy before the end of the 20th wk of gestation (1). Recurrent pregnancy loss (RPL) is defined as at least 2 consecutive miscarriages (2-4). It is estimated that 1-5% of couples suffer from RPL (5, 6). Although in 50% of cases several factors such as endocrine dysfunction, infections, environmental factors, and parental chromosomal abnormalities may cause RPL, in 50% of cases the cause is unknown (7-9).
Several studies have examined various polymorphisms of candidate genes that encode different mediators which may affect susceptibility to idiopathic RPL (4, 10, 11). A group of these genes belongs to metabolic enzymes. Genetic polymorphisms of these genes may affect the balance of phase I / phase II detoxification enzymes (12). One of these enzymes is encoded by CYP1A1 which is located on 15q24.1 and includes 7 exons. This gene acts in a 2-step process of detoxifying toxins. In the first step, CYP1A1 is required for activation of toxic components. These components are required for the 2nd detoxification step. The polymorphisms of this gene can be directly linked with functional disturbance diseases and conditions like cancers and idiopathic male infertility. A recent study showed that this gene can influence normal estrogen metabolism and placental function (13). It seems that its polymorphisms may also lead to RPL.
Another gene of this pathway is CYP2D6 which is located on 22q13.2 and includes 9 exons (14, 15). This gene is important for pharmacogenetics; it is involved in the metabolism of over 150 drugs and has been studied in individuals with suicidal thoughts or depression (16). CYP2D6 has an important role in catalyzing the oxidation of testosterone to androstenediones (17), both of which increase during pregnancy. So, it seems that any change in this enzyme may lead to an increased risk of RPL.
There is no autonomic innervation in fetoplacental blood vessels and the regulation of vascular functions at the fetomaternal interface is mediated by endothelial nitric oxide synthase (NOS3), which is a vasoactive mediator (18). This gene is located on 17q36.1 and includes 26 exons. It encodes an enzyme that is important in producing vascular NO (19, 20). "NO is a gaseous molecule, which serves different physiological regulatory functions in the regulation of reproduction, such as the formation of new blood vessels, enhancement of blood supply through the maternal arteries to the placenta, regulation of the placental vessel tones, and immune protection of the fetus. All these factors are required for a successful pregnancy outcome and any disturbance in these steps may increase the risk of miscarriage" (21). In the first trimester of pregnancy, trophoblast cells express a large amount of NOS3, so any polymorphism in coding or noncoding regions of the gene may change its activity or expression level, which may lead to RPL (19).
The aim of this study was to determine the relationship between 3 polymorphisms of these genes and RPL in the Iranian Azeri population.
2. Materials and Methods
2.1. Sampling
This case-control study was carried out in the Rahmani genetic lab, Tabriz, Iran, during April 2018-April 2020 with 2 groups. The control group (n = 136) consisted of 19-45 yr-old women with no history of miscarriage or infertility and with at least 1 successful pregnancy and a delivery without any complications. The case group (n = 136) consisted of 16-42 yr-old women with at least 2 consecutive idiopathic miscarriages and no successful pregnancies. It should also be considered that all the women in both groups were Iranians with Azeri origin.
All women with RPL due to infections, uterine conformational abnormalities, immune disorders, hormonal abnormalities (including thyroid and prolactin disorders) and chromosomal abnormalities in themselves or their spouses were excluded from the study.
The sample size was calculated with this formula: N = (Z1-α /2) 2 p (1-p) /d2, (Z1-α /2 = 1.96, p = 0.5, d = 0.1), N = 96. So at least 96 samples were needed to perform our study but in order to control random sampling the errors, we increased the sample size to 136 in each group. Please refer to table I for the demographic and clinical characteristics of the cases with RPL and the control group.
2.2. DNA extraction
5cc peripheral blood samples were taken from each individual. Genomic DNA was isolated from a 1 ml ethylenediamine tetraacetic acid-anticoagulated peripheral blood sample using a DNA extraction kit (KBC blood DNA extraction kit, Cat. No. K1135, Tehran, Iran) according to the manufacturers’ instructions. The quality of each sample was then estimated using a nanodrop for assessment of nucleic acid purity.
2.3. Genotyping
To confirm each polymorphism in its genetic region, the minor allele frequency (MAF) should be more than 0.01. Each single nucleotide polymorphism (SNP) was chosen according to its MAF in the 1000 Genome Project. MAF has been reported as C = 0.133387/668 for rs1048943, T = 0.063498/318 for rs28371725 and T = 0.361821/1812 for rs7830 (22).
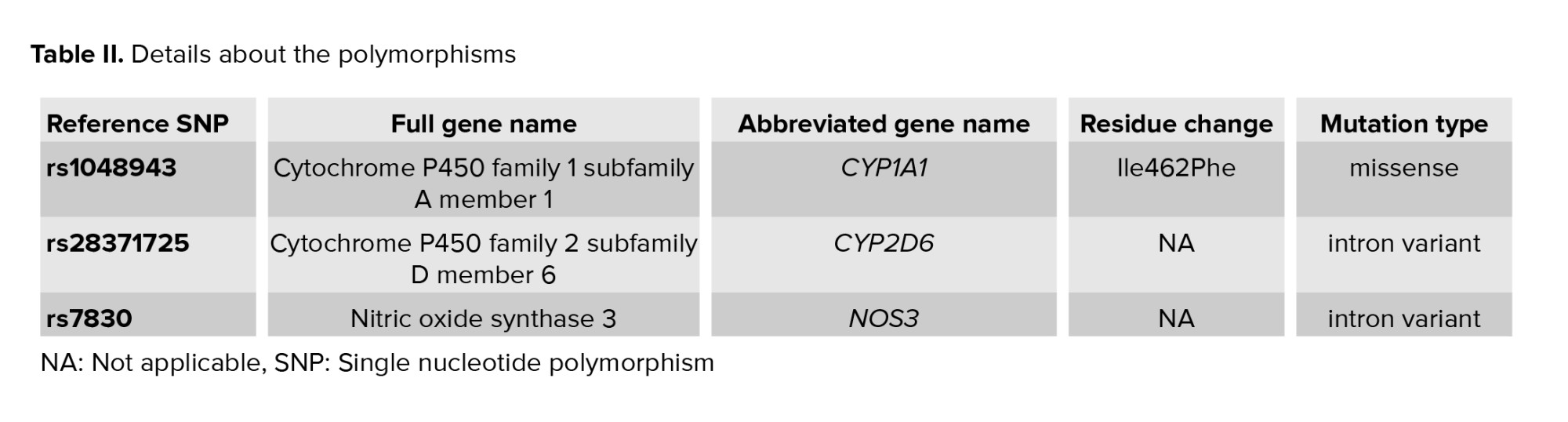
Rs1048943 is a missense coding sequence variant that causes Ile to Val change in the cyp1a1 protein; rs28371725 is a G>A intron variant which causes alternative splicing that can alter cyp2d6 function of the protein; and rs7830 is a G>T intron variant that leads to a different transcript. Detailed information about the polymorphisms including the full and abbreviated gene names, residue change and mutation type is presented in table ІІ.
Genetic polymorphisms of each participant for rs1048943 and rs28371725 were detected by allele-specific polymerase chain reaction (ARMS-PCR) and the polymorphisms for rs7830 were determined by tetra primer amplification refractory mutation system PCR (T-ARMS-PCR). The ARMS primers were designed using a WASP webpage with the address https://bioinfo.biotec.or.th/WASP/ and for T-ARMS-PCR the designs were done by the primer1 webpage with the address https://http://primer1.soton.ac.uk/. Detailed information about the primers is listed in table ІІІ and table ІV.
After their design, all the primers were evaluated with the BLAST-NCBI database and analyzed using the Oligo Analyzer software to ensure the validation of each primer. To increase the specificity of each forward primer in ARMS-PCR and each of the 2 inner primers in T-ARMS-PCR, an extra mismatch was designed in the 2nd nucleotide for ARMS-PCR and in the third nucleotide for T-ARMS-PCR from the 3′end.
Amplifications were carried out in a thermal cycler (Peqlab peqSTAR 96, Erlangen, Germany) with 2 tubes per person using ARMS-PCR, (1 for the wild allele and the other for the mutant allele) with a pair of primers in each tube (wild/mutant forward and common reverse) and 1 tube for each sample using T-ARMS-PCR with 4 primers in each tube. Each tube contained 50 ml: 25 ml of 2×PCRBIO Taq Mix Red Master Mix (Cat# PB10.13-02, London, England), 2.0 ml of each primer, 100-500 ng of genomic DNA, and up to 50 ml final volume of deionized water. After pre-denaturation at 95°C for 5 min, the PCR was carried out for 35 cycles of 30 sec at 94°C; 30 sec at 57°C for CYP1A1, 52°C for CYP2D6 and 69.3°C for NOS3; 30 sec at 72°C and at the end of the 35 cycles, the final extension at 72°C for 2 min to complete the extension of all DNA fragments. The PCR products were analyzed in 2% agarose gel stained with gel stain and with a 50 base pairs (bp) DNA ladder as the template of measurement. To ensure the accuracy of the SNP genotypes, 15% of the samples were selected randomly and re-genotyped to verify the initial results. The results confirmed that the genotyping was valid and consistent.
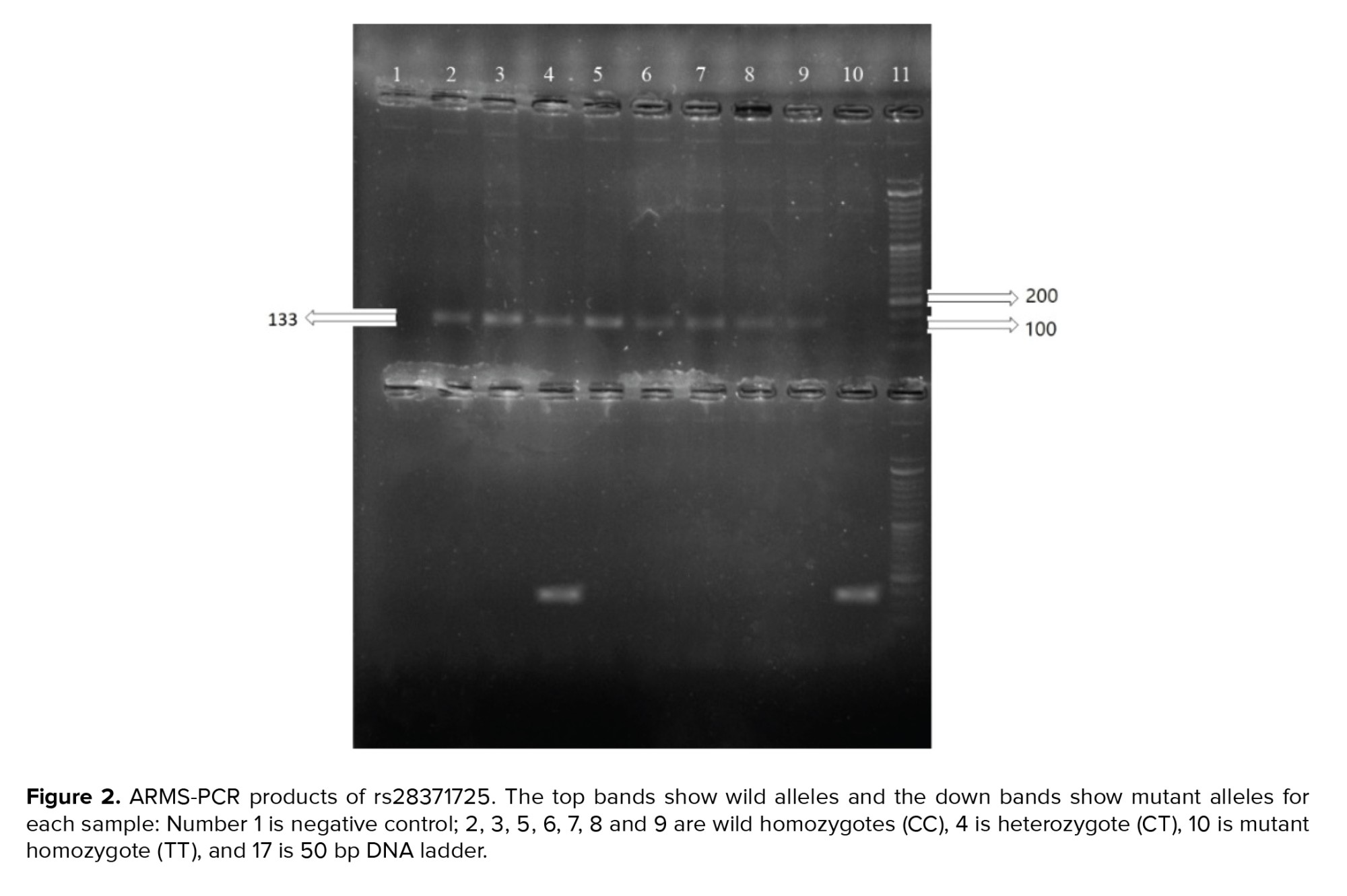
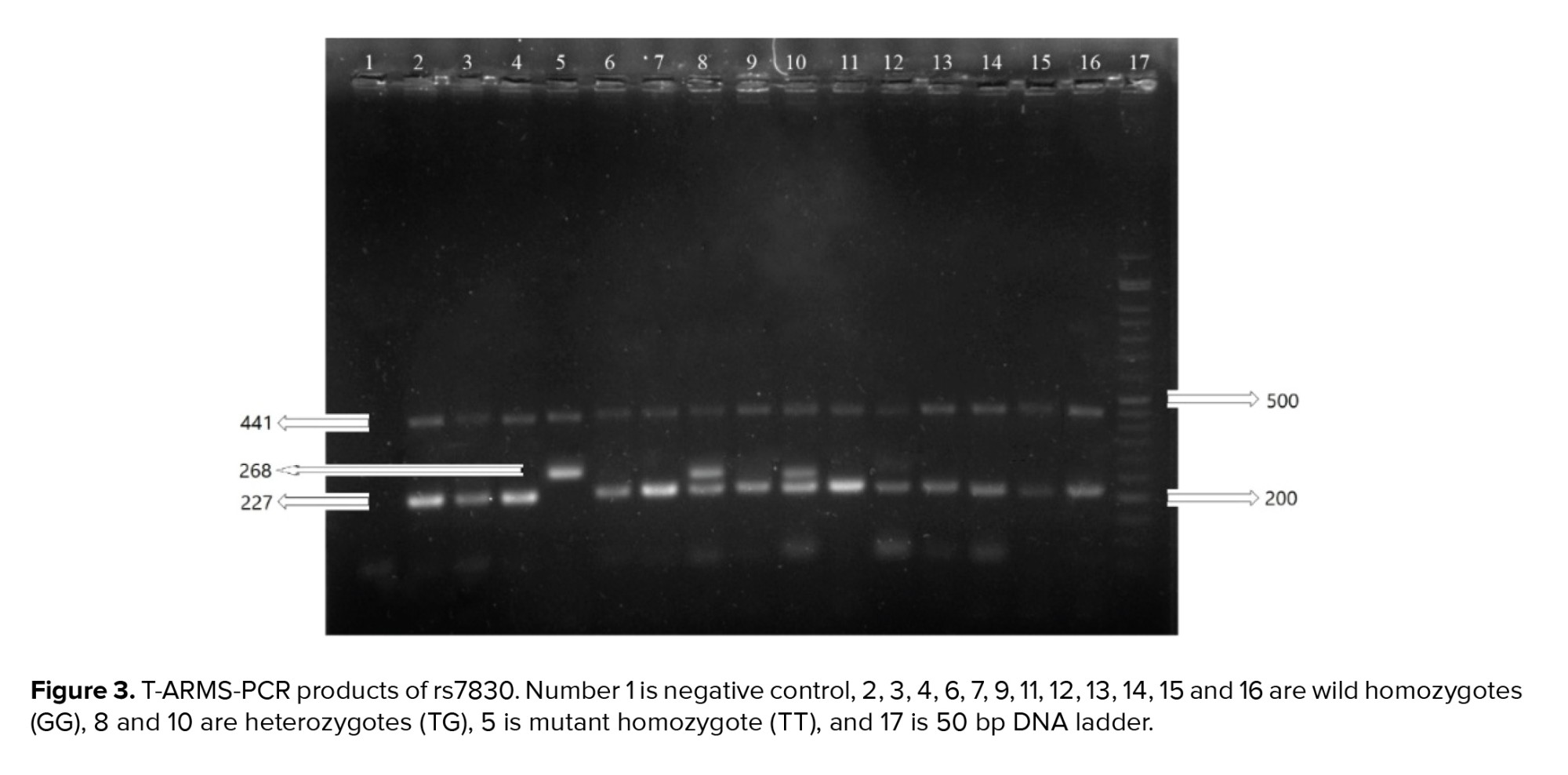
The sizes of the PCR products were 220 bp and 133 bp for the rs1048943 and rs28371725 polymorphisms, respectively, and for the rs7830 polymorphism the sizes were 441, 227 and 268 bp for the common, wild and mutant products, respectively. The PCR products are shown in figures 1, 2, and 3.




2.4. Ethical considerations
The Islamic Azad University, Tabriz Branch Ethics Committee, Tabriz, Iran approved this study (Code: IR.IAU.TABRIZ.REC.1398.022). All the participants completed a written informed consent form. The methods were performed in accordance with the ethical principles, national norms, standards, relevant guidelines, and regulations for conducting medical research in Iran.
2.5. Statistical analysis
Central tendency and dispersion of the demographic data related to the clinical characteristics of our case and control groups were examined by descriptive statistics such as mean and standard deviation.
Fisher’s exact test was used to compare the case and control groups. A p ˂ 0.05 was considered statistically significant. For the SNPs with a p ˂ 0.05, the Hardy-Weinberg equilibrium analysis was performed by using the Chi-square test and Fisher’s exact test (for multiplicative and additive models of rs1048943) to compare the observed and expected genotype frequencies in both the case and control groups to determine the model of association. It was found that the polymorphisms of all of the SNPs followed the multiplicative model. The additive model which is independent of the Hardy-Weinberg equilibrium was evaluated too. To show the effect of these polymorphisms on RPL, odds ratios (OR) with a 95% confidence interval (95% CI) were calculated using logistic regression, Chi-square test and Fisher’s exact test (for multiplicative and additive models of rs1048943) in the Statistical Package for the Social Sciences (SPSS), version 26 (SPSS Inc., Chicago, Illinois, USA).
3. Results
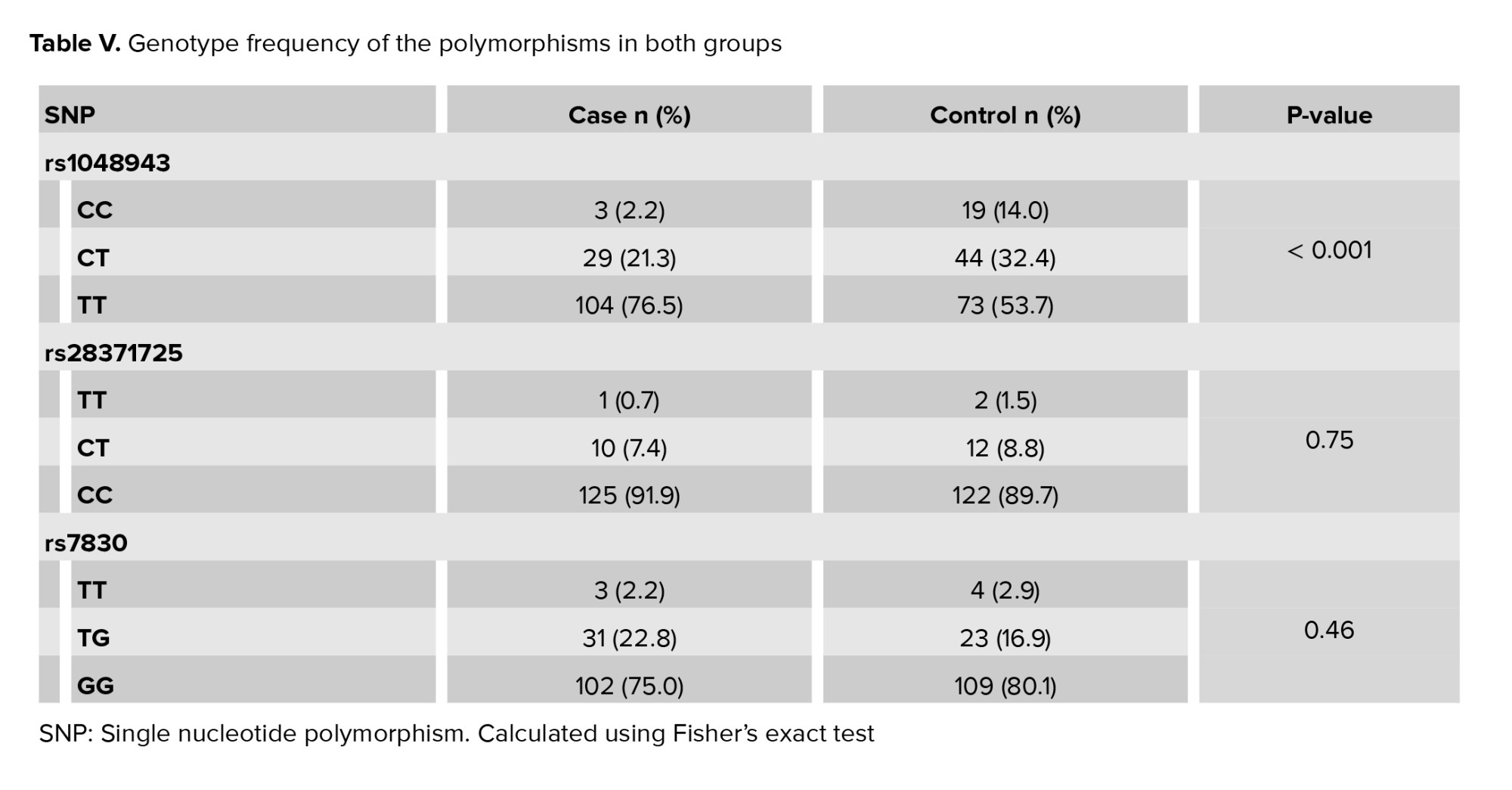
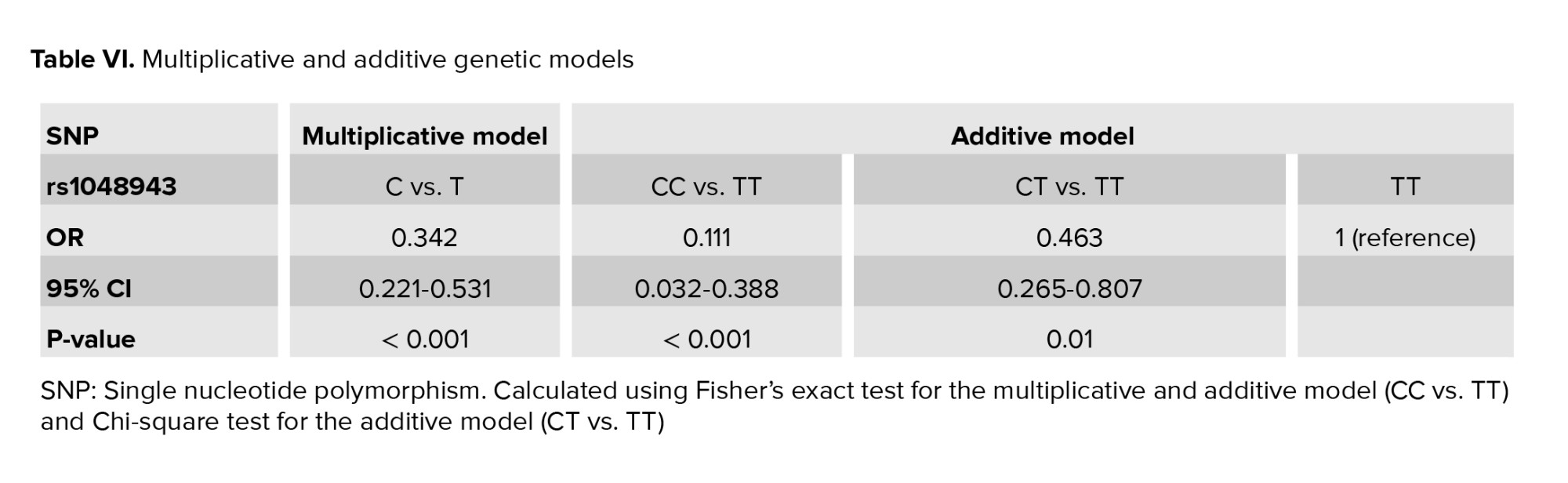
The mean age of the women in the control group was 31.9 ± 6.3 yr (range 19-45) and 28.9 ± 6.3 (range 16-42) in the case group. 3 candidate polymorphisms were genotyped and analyzed during our study. A comparison of the frequencies of the polymorphisms in the case and control groups are shown in table V. According to the results, rs28372725 of CYP2D6 (p = 0.75) and rs7830 of NOS3 (p = 0.46) did not show any statistically significant difference between the case and control groups and for this reason no more statistical steps were followed for these 2 SNPs. Regression analyses for rs1048943 of CYP1A1 (p < 0.001) under the multiplicative model and additive model were carried out and the results are shown in table VI. According to our findings, rs1048943 of CYP1A1 was the only polymorphism that was statistically associated with RPL in both multiplicative and additive models. As shown in table VI, the association of rs1048943 with RPL (p < 0.001, OR [95% CI] = 0.342 [0.221-0.531]) was observed under the multiplicative genetic model. In this SNP, TT is the wild genotype, so it was considered as the reference genotype. According to the additive genetic model, CC (OR [95% CI] = 0.111 [0.032-0.388], p < 0.001) and CT (OR [95% CI] = 0.463 [0.265-0.807], p = 0.01) of this SNP were associated with RPL using TT as the reference genotype.
Although the genotype frequencies of TT and CT in rs28372725 of CYP2D6 were higher in the controls than in the cases, we failed to find any statistically significant association of these with RPL. Finally, rs7830 of NOS3 did not show any significant association with RPL either.
4. Discussion
CYP1A1 plays an important role in the oxidation of polycyclic aromatic hydrocarbons like benzopyrene and polychlorinated biphenyls. These substances are unusual environmental toxicants (13). The role of this enzyme in activating carcinogens in cancers has been shown in previous studies (23-25). A recent study also showed that placental CYP1A1 mRNA levels were higher in women with RPL (26). We also know that CYP1A1 can affect the metabolism of estrogen and normal functions of the placenta (13). In fact, this enzyme participates in the metabolism of estrogen by catalyzing the 2-hydroxylation of stradiol (27) and can convert endogenous estrogens into more hydrophilic compounds (28). This enzyme also has an important role in the metabolism of enzymatic xenobiotics that are specifically induced in women exposed to tobacco smoke. These xenobiotics may transfer through the placenta to the fetus and cause toxicity in the fetus or may affect the expression or production of placental hormones and change the function of proteins (29). Thus, it seems that it could be a good candidate gene in relation to RPL.
One of its polymorphisms is rs1048943 A>G which is located in 2455 nucleotides and causes Ile>Val in the amino acid chain. This amino acid change occurs near the hem group of the protein and causes the enzyme activity to double (30). This polymorphism is a risk factor for pharyngeal, prostate, lung, oral, ovarian, bladder, colorectal, and cervical cancers (31). A protective association also was shown between this polymorphism and coronary artery disease in a study conducted with the Han population of China in 2017 (32). In our study, we found that rs1048943 was associated with a reduced risk of RPL in women of Iranian Azeri origin. However, another study done in 2014 in Russia did not show any association between this polymorphism and RPL (33). Given the varied results, studying this common polymorphism of CYP1A1 in larger populations with different genetic backgrounds is recommended.
CYP2D6 is active in catalyzing the oxidation of testosterone to androstenediones (17). Both of these hormones increase during pregnancy. The normal fertility process requires an adequate supply of sex hormones and the normal functioning of CYP2D6 is needed to reach this threshold. CYP2D6 is also an important modulator of the detoxification system, and can protect the environment of the utero-placental tissue from being affected by overwhelming oxidative stress (6). The activity of this enzyme begins to increase in the early stages of the 2nd trimester of pregnancy and continues to increase as the pregnancy progresses. Nowadays it is also known that many commonly used drugs in pregnancy are metabolized by this enzyme. Considering the highly polymorphic nature of this gene (28), it seems that this could be a good candidate gene in RPL associated studies. This enzyme is also important in pharmacogenetics and is active in the biotransformation of more than 150 drugs including antipsychotics, antidepressants, analgesics, beta-blockers, and antiarrhythmics (16).
Rs28371725 causes an A > G change in nucleotide 2989 in intron 6 which brings about alternative splicing and so changes the structure of its protein. This structure change results in a reduction in enzyme activity (reduction of metabolizing of beta-blockers). The influence of this polymorphism has been studied in cases of early onset of severe pre-eclampsia; its therapeutic responses and the association between this polymorphism and early-onset preeclampsia were observed (34). To the best of our knowledge, ours is the first study to evaluate the effect of this SNP in RPL. Our research showed that the association between this SNP and RPL in the Iranian Azeri population was not significant. Another study which was conducted in the South of India in 2004 investigated the association between rs3892097 of this gene and RPL but it also did not find any positive or negative association between rs3892097 and RPL (35).
"NOS3 encodes an enzyme that generates NO in endothelial cells and is involved in the regulation of vascular functions". The endometrial expression of NOS3 reaches its peak in humans during implantation (36). NO also plays an important role in blood pressure control, cardiovascular homeostasis, the metabolism of glucose, and insulin resistance during pregnancy. A reduction of NO production may cause pregnancy-related vascular problems like RPL. Additionally, alteration in NO synthesis may cause premature labor by affecting the inflammatory response and uterine contractility regulation (37). As a vasoactive agent, NO causes vasodilation and increases the rate of nutrient supply and oxygen perfusion in the umbilical cord. So, any decrease in its rate of production may cause intraurine hypoxia, restriction in fetal growth and increased feto-placental vascular resistance (38). We also know that the inhibition of NO synthesis in pregnant rats causes hypertension, proteinuria, thrombocytopenia and fetal growth restriction that can all lead to miscarriage (39). Hence, this gene could also be a good candidate gene for RPL associated studies.
Rs7830 (G>T) is associated with 2 different genes: NOS3 and ATG9B, but because it is located within a silencer motif, TGGGGAC, where a G>T change can lead to a different transcript, it seems that it influences the NOS3 gene more. Although we did not find any association between this SNP and RPL, the association of this SNP with end-stage renal disease has been previously demonstrated (40). The association between NOS polymorphisms and renal dysfunction (41), atherosclerotic vascular diseases (42), and advanced diabetic nephropathy (43) have also been shown. A study done in 2013 in China could not find any association between this SNP and RPL (44). Another study conducted in Russia in 2019 showed a positive association between rs2070744 of the NOS3 gene with miscarriage (20).
This research had some limitations. Firstly, we did not have any information about the women’s family history of miscarriage. Secondly, although we asked about the practice of smoking, we did not have any information about other lifestyle habits like alcohol consumption that may have affected the results. Thirdly, our study was limited to only 1 origin within 1 country as we did not assess RPL cases and healthy populations from other ethnic groups or countries. Fourthly, our case and control groups were relatively small, and finally, we investigated only 1 SNP from each gene. Therefore, in future studies, it would be beneficial to use larger groups from various nations, with awareness of the family history of miscarriage and lifestyle practices, and also to investigate other SNPs from these genes.
5. Conclusion
Our results indicated that while rs1048943 of CYP1A1 was associated with a decreased risk of RPL in the studied population, there was no statistically significant association between rs28372725 of CYP2D6 or rs7830 of NOS3 and RPL in the same population. Further studies in various populations are needed to confirm whether these polymorphisms have any positive or negative associations with RPL.
Acknowledgments
The authors would like to thank all the people in the control and patient groups for providing blood samples for the scientific research. This study did not receive any financial support.
Conflict of Interest
The authors declare that there is no conflict of interest.
Full-Text: (159 Views)
1. Introduction
Pregnancy loss is defined as a loss of pregnancy before the end of the 20th wk of gestation (1). Recurrent pregnancy loss (RPL) is defined as at least 2 consecutive miscarriages (2-4). It is estimated that 1-5% of couples suffer from RPL (5, 6). Although in 50% of cases several factors such as endocrine dysfunction, infections, environmental factors, and parental chromosomal abnormalities may cause RPL, in 50% of cases the cause is unknown (7-9).
Several studies have examined various polymorphisms of candidate genes that encode different mediators which may affect susceptibility to idiopathic RPL (4, 10, 11). A group of these genes belongs to metabolic enzymes. Genetic polymorphisms of these genes may affect the balance of phase I / phase II detoxification enzymes (12). One of these enzymes is encoded by CYP1A1 which is located on 15q24.1 and includes 7 exons. This gene acts in a 2-step process of detoxifying toxins. In the first step, CYP1A1 is required for activation of toxic components. These components are required for the 2nd detoxification step. The polymorphisms of this gene can be directly linked with functional disturbance diseases and conditions like cancers and idiopathic male infertility. A recent study showed that this gene can influence normal estrogen metabolism and placental function (13). It seems that its polymorphisms may also lead to RPL.
Another gene of this pathway is CYP2D6 which is located on 22q13.2 and includes 9 exons (14, 15). This gene is important for pharmacogenetics; it is involved in the metabolism of over 150 drugs and has been studied in individuals with suicidal thoughts or depression (16). CYP2D6 has an important role in catalyzing the oxidation of testosterone to androstenediones (17), both of which increase during pregnancy. So, it seems that any change in this enzyme may lead to an increased risk of RPL.
There is no autonomic innervation in fetoplacental blood vessels and the regulation of vascular functions at the fetomaternal interface is mediated by endothelial nitric oxide synthase (NOS3), which is a vasoactive mediator (18). This gene is located on 17q36.1 and includes 26 exons. It encodes an enzyme that is important in producing vascular NO (19, 20). "NO is a gaseous molecule, which serves different physiological regulatory functions in the regulation of reproduction, such as the formation of new blood vessels, enhancement of blood supply through the maternal arteries to the placenta, regulation of the placental vessel tones, and immune protection of the fetus. All these factors are required for a successful pregnancy outcome and any disturbance in these steps may increase the risk of miscarriage" (21). In the first trimester of pregnancy, trophoblast cells express a large amount of NOS3, so any polymorphism in coding or noncoding regions of the gene may change its activity or expression level, which may lead to RPL (19).
The aim of this study was to determine the relationship between 3 polymorphisms of these genes and RPL in the Iranian Azeri population.
2. Materials and Methods
2.1. Sampling
This case-control study was carried out in the Rahmani genetic lab, Tabriz, Iran, during April 2018-April 2020 with 2 groups. The control group (n = 136) consisted of 19-45 yr-old women with no history of miscarriage or infertility and with at least 1 successful pregnancy and a delivery without any complications. The case group (n = 136) consisted of 16-42 yr-old women with at least 2 consecutive idiopathic miscarriages and no successful pregnancies. It should also be considered that all the women in both groups were Iranians with Azeri origin.
All women with RPL due to infections, uterine conformational abnormalities, immune disorders, hormonal abnormalities (including thyroid and prolactin disorders) and chromosomal abnormalities in themselves or their spouses were excluded from the study.
The sample size was calculated with this formula: N = (Z1-α /2) 2 p (1-p) /d2, (Z1-α /2 = 1.96, p = 0.5, d = 0.1), N = 96. So at least 96 samples were needed to perform our study but in order to control random sampling the errors, we increased the sample size to 136 in each group. Please refer to table I for the demographic and clinical characteristics of the cases with RPL and the control group.
2.2. DNA extraction
5cc peripheral blood samples were taken from each individual. Genomic DNA was isolated from a 1 ml ethylenediamine tetraacetic acid-anticoagulated peripheral blood sample using a DNA extraction kit (KBC blood DNA extraction kit, Cat. No. K1135, Tehran, Iran) according to the manufacturers’ instructions. The quality of each sample was then estimated using a nanodrop for assessment of nucleic acid purity.
2.3. Genotyping
To confirm each polymorphism in its genetic region, the minor allele frequency (MAF) should be more than 0.01. Each single nucleotide polymorphism (SNP) was chosen according to its MAF in the 1000 Genome Project. MAF has been reported as C = 0.133387/668 for rs1048943, T = 0.063498/318 for rs28371725 and T = 0.361821/1812 for rs7830 (22).
Rs1048943 is a missense coding sequence variant that causes Ile to Val change in the cyp1a1 protein; rs28371725 is a G>A intron variant which causes alternative splicing that can alter cyp2d6 function of the protein; and rs7830 is a G>T intron variant that leads to a different transcript. Detailed information about the polymorphisms including the full and abbreviated gene names, residue change and mutation type is presented in table ІІ.
Genetic polymorphisms of each participant for rs1048943 and rs28371725 were detected by allele-specific polymerase chain reaction (ARMS-PCR) and the polymorphisms for rs7830 were determined by tetra primer amplification refractory mutation system PCR (T-ARMS-PCR). The ARMS primers were designed using a WASP webpage with the address https://bioinfo.biotec.or.th/WASP/ and for T-ARMS-PCR the designs were done by the primer1 webpage with the address https://http://primer1.soton.ac.uk/. Detailed information about the primers is listed in table ІІІ and table ІV.
After their design, all the primers were evaluated with the BLAST-NCBI database and analyzed using the Oligo Analyzer software to ensure the validation of each primer. To increase the specificity of each forward primer in ARMS-PCR and each of the 2 inner primers in T-ARMS-PCR, an extra mismatch was designed in the 2nd nucleotide for ARMS-PCR and in the third nucleotide for T-ARMS-PCR from the 3′end.
Amplifications were carried out in a thermal cycler (Peqlab peqSTAR 96, Erlangen, Germany) with 2 tubes per person using ARMS-PCR, (1 for the wild allele and the other for the mutant allele) with a pair of primers in each tube (wild/mutant forward and common reverse) and 1 tube for each sample using T-ARMS-PCR with 4 primers in each tube. Each tube contained 50 ml: 25 ml of 2×PCRBIO Taq Mix Red Master Mix (Cat# PB10.13-02, London, England), 2.0 ml of each primer, 100-500 ng of genomic DNA, and up to 50 ml final volume of deionized water. After pre-denaturation at 95°C for 5 min, the PCR was carried out for 35 cycles of 30 sec at 94°C; 30 sec at 57°C for CYP1A1, 52°C for CYP2D6 and 69.3°C for NOS3; 30 sec at 72°C and at the end of the 35 cycles, the final extension at 72°C for 2 min to complete the extension of all DNA fragments. The PCR products were analyzed in 2% agarose gel stained with gel stain and with a 50 base pairs (bp) DNA ladder as the template of measurement. To ensure the accuracy of the SNP genotypes, 15% of the samples were selected randomly and re-genotyped to verify the initial results. The results confirmed that the genotyping was valid and consistent.
The sizes of the PCR products were 220 bp and 133 bp for the rs1048943 and rs28371725 polymorphisms, respectively, and for the rs7830 polymorphism the sizes were 441, 227 and 268 bp for the common, wild and mutant products, respectively. The PCR products are shown in figures 1, 2, and 3.







2.4. Ethical considerations
The Islamic Azad University, Tabriz Branch Ethics Committee, Tabriz, Iran approved this study (Code: IR.IAU.TABRIZ.REC.1398.022). All the participants completed a written informed consent form. The methods were performed in accordance with the ethical principles, national norms, standards, relevant guidelines, and regulations for conducting medical research in Iran.
2.5. Statistical analysis
Central tendency and dispersion of the demographic data related to the clinical characteristics of our case and control groups were examined by descriptive statistics such as mean and standard deviation.
Fisher’s exact test was used to compare the case and control groups. A p ˂ 0.05 was considered statistically significant. For the SNPs with a p ˂ 0.05, the Hardy-Weinberg equilibrium analysis was performed by using the Chi-square test and Fisher’s exact test (for multiplicative and additive models of rs1048943) to compare the observed and expected genotype frequencies in both the case and control groups to determine the model of association. It was found that the polymorphisms of all of the SNPs followed the multiplicative model. The additive model which is independent of the Hardy-Weinberg equilibrium was evaluated too. To show the effect of these polymorphisms on RPL, odds ratios (OR) with a 95% confidence interval (95% CI) were calculated using logistic regression, Chi-square test and Fisher’s exact test (for multiplicative and additive models of rs1048943) in the Statistical Package for the Social Sciences (SPSS), version 26 (SPSS Inc., Chicago, Illinois, USA).
3. Results
The mean age of the women in the control group was 31.9 ± 6.3 yr (range 19-45) and 28.9 ± 6.3 (range 16-42) in the case group. 3 candidate polymorphisms were genotyped and analyzed during our study. A comparison of the frequencies of the polymorphisms in the case and control groups are shown in table V. According to the results, rs28372725 of CYP2D6 (p = 0.75) and rs7830 of NOS3 (p = 0.46) did not show any statistically significant difference between the case and control groups and for this reason no more statistical steps were followed for these 2 SNPs. Regression analyses for rs1048943 of CYP1A1 (p < 0.001) under the multiplicative model and additive model were carried out and the results are shown in table VI. According to our findings, rs1048943 of CYP1A1 was the only polymorphism that was statistically associated with RPL in both multiplicative and additive models. As shown in table VI, the association of rs1048943 with RPL (p < 0.001, OR [95% CI] = 0.342 [0.221-0.531]) was observed under the multiplicative genetic model. In this SNP, TT is the wild genotype, so it was considered as the reference genotype. According to the additive genetic model, CC (OR [95% CI] = 0.111 [0.032-0.388], p < 0.001) and CT (OR [95% CI] = 0.463 [0.265-0.807], p = 0.01) of this SNP were associated with RPL using TT as the reference genotype.
Although the genotype frequencies of TT and CT in rs28372725 of CYP2D6 were higher in the controls than in the cases, we failed to find any statistically significant association of these with RPL. Finally, rs7830 of NOS3 did not show any significant association with RPL either.


4. Discussion
CYP1A1 plays an important role in the oxidation of polycyclic aromatic hydrocarbons like benzopyrene and polychlorinated biphenyls. These substances are unusual environmental toxicants (13). The role of this enzyme in activating carcinogens in cancers has been shown in previous studies (23-25). A recent study also showed that placental CYP1A1 mRNA levels were higher in women with RPL (26). We also know that CYP1A1 can affect the metabolism of estrogen and normal functions of the placenta (13). In fact, this enzyme participates in the metabolism of estrogen by catalyzing the 2-hydroxylation of stradiol (27) and can convert endogenous estrogens into more hydrophilic compounds (28). This enzyme also has an important role in the metabolism of enzymatic xenobiotics that are specifically induced in women exposed to tobacco smoke. These xenobiotics may transfer through the placenta to the fetus and cause toxicity in the fetus or may affect the expression or production of placental hormones and change the function of proteins (29). Thus, it seems that it could be a good candidate gene in relation to RPL.
One of its polymorphisms is rs1048943 A>G which is located in 2455 nucleotides and causes Ile>Val in the amino acid chain. This amino acid change occurs near the hem group of the protein and causes the enzyme activity to double (30). This polymorphism is a risk factor for pharyngeal, prostate, lung, oral, ovarian, bladder, colorectal, and cervical cancers (31). A protective association also was shown between this polymorphism and coronary artery disease in a study conducted with the Han population of China in 2017 (32). In our study, we found that rs1048943 was associated with a reduced risk of RPL in women of Iranian Azeri origin. However, another study done in 2014 in Russia did not show any association between this polymorphism and RPL (33). Given the varied results, studying this common polymorphism of CYP1A1 in larger populations with different genetic backgrounds is recommended.
CYP2D6 is active in catalyzing the oxidation of testosterone to androstenediones (17). Both of these hormones increase during pregnancy. The normal fertility process requires an adequate supply of sex hormones and the normal functioning of CYP2D6 is needed to reach this threshold. CYP2D6 is also an important modulator of the detoxification system, and can protect the environment of the utero-placental tissue from being affected by overwhelming oxidative stress (6). The activity of this enzyme begins to increase in the early stages of the 2nd trimester of pregnancy and continues to increase as the pregnancy progresses. Nowadays it is also known that many commonly used drugs in pregnancy are metabolized by this enzyme. Considering the highly polymorphic nature of this gene (28), it seems that this could be a good candidate gene in RPL associated studies. This enzyme is also important in pharmacogenetics and is active in the biotransformation of more than 150 drugs including antipsychotics, antidepressants, analgesics, beta-blockers, and antiarrhythmics (16).
Rs28371725 causes an A > G change in nucleotide 2989 in intron 6 which brings about alternative splicing and so changes the structure of its protein. This structure change results in a reduction in enzyme activity (reduction of metabolizing of beta-blockers). The influence of this polymorphism has been studied in cases of early onset of severe pre-eclampsia; its therapeutic responses and the association between this polymorphism and early-onset preeclampsia were observed (34). To the best of our knowledge, ours is the first study to evaluate the effect of this SNP in RPL. Our research showed that the association between this SNP and RPL in the Iranian Azeri population was not significant. Another study which was conducted in the South of India in 2004 investigated the association between rs3892097 of this gene and RPL but it also did not find any positive or negative association between rs3892097 and RPL (35).
"NOS3 encodes an enzyme that generates NO in endothelial cells and is involved in the regulation of vascular functions". The endometrial expression of NOS3 reaches its peak in humans during implantation (36). NO also plays an important role in blood pressure control, cardiovascular homeostasis, the metabolism of glucose, and insulin resistance during pregnancy. A reduction of NO production may cause pregnancy-related vascular problems like RPL. Additionally, alteration in NO synthesis may cause premature labor by affecting the inflammatory response and uterine contractility regulation (37). As a vasoactive agent, NO causes vasodilation and increases the rate of nutrient supply and oxygen perfusion in the umbilical cord. So, any decrease in its rate of production may cause intraurine hypoxia, restriction in fetal growth and increased feto-placental vascular resistance (38). We also know that the inhibition of NO synthesis in pregnant rats causes hypertension, proteinuria, thrombocytopenia and fetal growth restriction that can all lead to miscarriage (39). Hence, this gene could also be a good candidate gene for RPL associated studies.
Rs7830 (G>T) is associated with 2 different genes: NOS3 and ATG9B, but because it is located within a silencer motif, TGGGGAC, where a G>T change can lead to a different transcript, it seems that it influences the NOS3 gene more. Although we did not find any association between this SNP and RPL, the association of this SNP with end-stage renal disease has been previously demonstrated (40). The association between NOS polymorphisms and renal dysfunction (41), atherosclerotic vascular diseases (42), and advanced diabetic nephropathy (43) have also been shown. A study done in 2013 in China could not find any association between this SNP and RPL (44). Another study conducted in Russia in 2019 showed a positive association between rs2070744 of the NOS3 gene with miscarriage (20).
This research had some limitations. Firstly, we did not have any information about the women’s family history of miscarriage. Secondly, although we asked about the practice of smoking, we did not have any information about other lifestyle habits like alcohol consumption that may have affected the results. Thirdly, our study was limited to only 1 origin within 1 country as we did not assess RPL cases and healthy populations from other ethnic groups or countries. Fourthly, our case and control groups were relatively small, and finally, we investigated only 1 SNP from each gene. Therefore, in future studies, it would be beneficial to use larger groups from various nations, with awareness of the family history of miscarriage and lifestyle practices, and also to investigate other SNPs from these genes.
5. Conclusion
Our results indicated that while rs1048943 of CYP1A1 was associated with a decreased risk of RPL in the studied population, there was no statistically significant association between rs28372725 of CYP2D6 or rs7830 of NOS3 and RPL in the same population. Further studies in various populations are needed to confirm whether these polymorphisms have any positive or negative associations with RPL.
Acknowledgments
The authors would like to thank all the people in the control and patient groups for providing blood samples for the scientific research. This study did not receive any financial support.
Conflict of Interest
The authors declare that there is no conflict of interest.
Type of Study: Original Article |
Subject:
Reproductive Genetics
References
1. Diaz-Nunez M, Rabanal A, Exposito A, Ferrando M, Quintana F, Soria JM, et al. Recurrent miscarriage and implantation failure of unknown cause studied by a panel of thrombophilia conditions: Increased frequency of FXIII Val34Leu polymorphism. J Reprod Infertil 2019; 20: 76-82.
2. Asgari N, Akbari MT, Zare Sh, Babamohammadi Gh. Positive association of apolipoprotein E4 polymorphism with recurrent pregnancy loss in Iranian patients. J Assist Reprod Genet 2013; 30: 265-268. [DOI:10.1007/s10815-012-9897-5] [PMID] [PMCID]
3. Hashemi M, Mokhtari M, Khazaeian S, Bahari G, Rezaei M, Nakhaee A, et al. Evaluation of HLA-G 14-bp ins/del and +3142G>C polymorphisms with susceptibility to recurrent spontaneous abortion. Taiwan J Obstet Gynecol 2017; 56: 276-280. [DOI:10.1016/j.tjog.2017.04.002] [PMID]
4. Zhang M, Xu J, Bao X, Niu W, Wang L, Du L, et al. Association between genetic polymorphisms in interleukin genes and recurrent pregnancy loss: A systematic review and meta-analysis. PLoS One 2017; 12: e0169891. [DOI:10.1371/journal.pone.0169891] [PMID] [PMCID]
5. Karatas A, Eroz R, Albayrak M, Ozlu T, Cakmak B, Keskin F. Evaluation of chromosomal abnormalities and common trombophilic mutations in cases with recurrent miscarriage. Afr Health Sci 2014; 14: 216-222. [DOI:10.4314/ahs.v14i1.34] [PMID] [PMCID]
6. Shi X, Xie X, Jia Y, Li S. Maternal genetic polymorphisms and unexplained recurrent miscarriage: A systematic review and meta-analysis. Clin Genet 2017; 91: 265-284. [DOI:10.1111/cge.12910] [PMID]
7. Li J, Chen Y, Wu H, Li L. Apolipoprotein E (Apo E) gene polymorphisms and recurrent pregnancy loss: A meta-analysis. J Assist Reprod Genet 2014; 31: 139-148. [DOI:10.1007/s10815-013-0128-5] [PMID] [PMCID]
8. Pereza N, Ostojic S, Kapovic M, Peterlin B. Systematic review and meta-analysis of genetic association studies in idiopathic recurrent spontaneous abortion. Fertil Steril 2017; 107: 150-159. [DOI:10.1016/j.fertnstert.2016.10.007] [PMID]
9. Du B, Shi X, Yin C, Feng X. Polymorphisms of methalenetetrahydrofolate reductase in recurrent pregnancy loss: An overview of systematic reviews and meta-analyses. J Assist Reprod Genet 2019; 36: 1315-1328. [DOI:10.1007/s10815-019-01473-2] [PMID] [PMCID]
10. Poursadegh Zonouzi A, Chaparzadeh N, Ghorbian S, Sadaghiani MM, Farzadi L, Ghasemzadeh A, et al. The association between thrombophilic gene mutations and recurrent pregnancy loss. J Assist Reprod Genet 2013; 30: 1353-1359. [DOI:10.1007/s10815-013-0071-5] [PMID] [PMCID]
11. Tur-Torres MH, Garrido-Gimenez C, Alijotas-Reig J. Genetics of recurrent miscarriage and fetal loss. Best Pract Res Clin Obstet Gynaecol 2017; 42: 11-25. [DOI:10.1016/j.bpobgyn.2017.03.007] [PMID]
12. Zong Ch, Sha Y, Xiang H, Wang J, Chen D, Liu J, et al. Glutathione S-transferase A1 polymorphism and the risk of recurrent spontaneous abortion in Chinese Han population. J Assist Reprod Genet 2014; 31: 379-382. [DOI:10.1007/s10815-013-0163-2] [PMID] [PMCID]
13. Li J, Chen Y, Mo S, Nai D. Potential positive association between cytochrome P450 1A1 gene polymorphisms and recurrent pregnancy loss: A meta-analysis. Ann Hum Genet 2017; 81: 161-173. [DOI:10.1111/ahg.12196] [PMID]
14. Nofziger Ch, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agundez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther 2020; 107: 154-170. [DOI:10.1002/cpt.1643] [PMID] [PMCID]
15. Yang Y, Botton MR, Scott ER, Scott SA. Sequencing the CYP2D6 gene: From variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 2017; 18: 673-685. [DOI:10.2217/pgs-2017-0033] [PMID] [PMCID]
16. Skadric I, Stojkovic O. Defining screening panel of functional variants of CYP1A1, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 genes in Serbian population. Int J Legal Med 2020; 134: 433-439. [DOI:10.1007/s00414-019-02234-7] [PMID]
17. Monostory K, Dvorak Z. Steroid regulation of drug-metabolizing cytochromes P450. Curr Drug Metab 2011; 12: 154-172. [DOI:10.2174/138920011795016854] [PMID]
18. Pereza N, Peterlin B, Volk M, Kapovic M, Ostojic S. A critical update on endothelial nitric oxide synthase gene variations in women with idiopathic recurrent spontaneous abortion: Genetic association study, systematic review and meta-analyses. Mol Hum Reprod 2015; 21: 466-478. [DOI:10.1093/molehr/gav008] [PMID]
19. Parveen F, Faridi RM, Alam S, Agrawal S. Genetic analysis of eNOS gene polymorphisms in association with recurrent miscarriage among North Indian women. Reprod Biomed Online 2011; 23: 124-131. [DOI:10.1016/j.rbmo.2011.03.022] [PMID]
20. Trifonova EA, Swarovskaya MG, Ganzha OA, Voronkova OV, Gabidulina TV, Stepanov VA. The interaction effect of angiogenesis and endothelial dysfunction-related gene variants increases the susceptibility of recurrent pregnancy loss. J Assist Reprod Genet 2019; 36: 717-726. [DOI:10.1007/s10815-019-01403-2] [PMID] [PMCID]
21. Azani A, Hosseinzadeh A, Azadkhah R, Zonouzi AAP, Zonouzi AP, Aftabi Y, et al. Association of endothelial nitric oxide synthase gene variants (-786 T>C, intron 4 b/a VNTR and 894 G>T) with idiopathic recurrent pregnancy loss: A case-control study with haplotype and in silico analysis. Eur J Obstet Gynecol Reprod Biol 2017; 215: 93-100. [DOI:10.1016/j.ejogrb.2017.05.024] [PMID]
22. dbSNP. Available at: https://www.ncbi.nlm.nih.gov/snp.
23. Wu H, Ouyang Q, Tian C, Xie N, Cao M, Shui ZR. Cytochrome P450 1A1 (CYP1A1) Gene polymorphisms and susceptibility to breast cancer: A meta-analysis in the Chinese Population. Clin Lab 2017; 63: 67-72. [DOI:10.7754/Clin.Lab.2016.160535] [PMID]
24. Akhtar S, Mahjabeen I, Akram Z, Kayani MA. CYP1A1 and GSTP1 gene variations in breast cancer: A systematic review and case-control study. Fam Cancer 2016; 15: 201-214. [DOI:10.1007/s10689-015-9849-1] [PMID]
25. Mrozikiewicz PM, Grześkowiak E, Seremak-Mrozikiewicz A, Bogacz A, Barlik M, Semczuk A, et al. Importance of CYP1A1 polymorphism and its transcriptional regulation in ovarian and endometrial cancer. Ginekol Pol 2011; 82: 925-932.
26. Wu Y, Chen X, Chang X, Huang YJ, Bao S, He Q, et al. Potential involvement of placental AhR in unexplained recurrent spontaneous abortion. Reprod Toxicol 2016; 59: 45-52. [DOI:10.1016/j.reprotox.2015.11.005] [PMID]
27. de Oliveira CBM, Cardoso-Filho C, Bossi LS, Lourenço GJ, Costa-Gurgel MS, Lima CS. Association of CYP1A1 A4889G and T6235C polymorphisms with the risk of sporadic breast cancer in Brazilian women. Clinics 2015; 70: 680-685. [DOI:10.6061/clinics/2015(10)04]
28. Mao Y, Zhang K, Ma L, Yun X, Ou F, Liu G, et al. Interaction between CYP1A1/CYP17A1 polymorphisms and parental risk factors in the risk of hypospadias in a Chinese population. Sci Rep 2019; 9: 4123. [DOI:10.1038/s41598-019-40755-8] [PMID] [PMCID]
29. Mohammed AM, Huuskonen P, Juvonen R, Sahlman H, Repo J, Myöhänen K, et al. Activities of metabolizing enzymes in human placenta. Toxicol Lett 2020; 326: 70-77. [DOI:10.1016/j.toxlet.2020.02.014] [PMID]
30. Abbas M, Srivastava K, Imran M, Banerjee M. Association of CYP1A1 gene variants rs4646903 (T>C) and rs1048943 (A>G) with cervical cancer in a North Indian population. Eur J Obstet Gynecol Reprod Biol 2014; 176: 68-74. [DOI:10.1016/j.ejogrb.2014.02.036] [PMID]
31. Jain V, Ratre YK, Amle D, Mishra PK, Patra PK. Polymorphism of CYP1A1 gene variants rs4646903 and rs1048943 relation to the incidence of cervical cancer in Chhattisgarh. Environ Toxicol Pharmacol 2017; 52: 188-192. [DOI:10.1016/j.etap.2017.04.009] [PMID]
32. Peng DD, Xie W, Yu ZX. Impact of interaction between CYP1A1 genetic polymorphisms and smoking on coronary artery disease in the Han of China. Clin Exp Hypertens 2017; 39: 339-343. [DOI:10.1080/10641963.2016.1259326] [PMID]
33. Khadzhieva MB, Lutcenko NN, Volodin IV, Morozova KV, Salnikova LE. Association of oxidative stress-related genes with idiopathic recurrent miscarriage. Free Radic Res 2014; 48: 534-541. [DOI:10.3109/10715762.2014.891735] [PMID]
34. Sun ChJ, Li L, Li XY, Zhang WY, Liu XW. Associations of polymorphisms of CYP2D6 and CYP2C9 with early onset severe pre-eclampsia and response to labetalol therapy. Arch Gynecol Obstet 2018; 298: 125-132. [DOI:10.1007/s00404-018-4791-8] [PMID]
35. Suryanarayana V, Deenadayal M, Singh L. Association of CYP1A1 gene polymorphism with recurrent pregnancy loss in the South Indian population. Hum Reprod 2004; 19: 2648-2652. [DOI:10.1093/humrep/deh463] [PMID]
36. Najafi T, Ghaffari Novin M, Ghazi R, Khorram O. Altered endometrial expression of endothelial nitric oxide synthase in women with unexplained recurrent miscarriage and infertility. Reprod Biomed Online 2012; 25: 408-414. [DOI:10.1016/j.rbmo.2012.07.004] [PMID] [PMCID]
37. Barbitoff YA, Tsarev AA, Vashukova ES, Maksiutenko EM, Kovalenko LV, Belotserkovtseva LD, et al. A data-driven review of the genetic factors of pregnancy complications. Int J Mol Sci 2020; 21: 3384. [DOI:10.3390/ijms21093384] [PMID] [PMCID]
38. Chakraborty P, Khamit A, Hermesz E. Fetal oxygen supply can be improved by an effective cross-talk between fetal erythrocytes and vascular endothelium. Biochim Biophys Acta Mol Basis Dis 2021; 1867: 166243. [DOI:10.1016/j.bbadis.2021.166243] [PMID]
39. Al Sallout RJ, Sharif FA. Polymorphisms in NOS3, ACE and PAI-1 genes and risk of spontaneous recurrent miscarriage in the Gaza Strip. Med Princ Pract 2010; 19: 99-104. [DOI:10.1159/000273067] [PMID]
40. Jimenez-Sousa MA, Lopez E, Fernandez-Rodriguez A, Tamayo E, Fernandez-Navarro P, Segura-Roda L, et al. Genetic polymorphisms located in genes related to immune and inflammatory processes are associated with end-stage renal disease: A preliminary study. BMC Med Genet 2012; 13: 58. [DOI:10.1186/1471-2350-13-58] [PMID] [PMCID]
41. Popov AF, Hinz J, Schulz EG, Schmitto JD, Wiese CH, Quintel M, et al. The eNOS 786C/T polymorphism in cardiac surgical patients with cardiopulmonary bypass is associated with renal dysfunction. Eur J Cardiothorac Surg 2009; 36: 651-656. [DOI:10.1016/j.ejcts.2009.04.049] [PMID]
42. Kullo IJ, Greene MT, Boerwinkle E, Chu J, Turner ST, Kardia SL. Association of polymorphisms in NOS3 with the ankle-brachial index in hypertensive adults. Atherosclerosis 2008; 196: 905-912. [DOI:10.1016/j.atherosclerosis.2007.02.008] [PMID] [PMCID]
43. Wang ChH, Li F, Hiller S, Kim HS, Maeda N, Smithies O, et al. A modest decrease in endothelial NOS in mice comparable to that associated with human NOS3 variants exacerbates diabetic nephropathy. Proc Natl Acad Sci U S A 2011; 108: 2070-2075. [DOI:10.1073/pnas.1018766108] [PMID] [PMCID]
44. Luo L, Li DH, Wei SG, Zhang HB, Li SB, Zhao J. Polymorphisms in the endothelial nitric oxide synthase gene associated with recurrent miscarriage. Genet Mol Res 2013; 12: 3879-3886. [DOI:10.4238/2013.September.23.6] [PMID]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





