Sat, Feb 21, 2026
[Archive]
Volume 18, Issue 9 (September 2020)
IJRM 2020, 18(9): 747-754 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Ardeshir F, Keshavarz L, Asadian F, Omidmokhtarkhanloo G, Yavarian M. Role of the 820 A/G variant in the IGF-2 gene and recurrent spontaneous abortion in southern Iran: A cross-sectional study. IJRM 2020; 18 (9) :747-754
URL: http://ijrm.ir/article-1-1411-en.html
URL: http://ijrm.ir/article-1-1411-en.html
Farzaneh Ardeshir1 

 , Leila Keshavarz1
, Leila Keshavarz1 

 , Fatemeh Asadian2
, Fatemeh Asadian2 

 , Gohar Omidmokhtarkhanloo1
, Gohar Omidmokhtarkhanloo1 

 , Majid Yavarian *3
, Majid Yavarian *3 




 , Leila Keshavarz1
, Leila Keshavarz1 

 , Fatemeh Asadian2
, Fatemeh Asadian2 

 , Gohar Omidmokhtarkhanloo1
, Gohar Omidmokhtarkhanloo1 

 , Majid Yavarian *3
, Majid Yavarian *3 


1- Department of Biology, Islamic Azad University, Arsanjan Branch, Arsanjan, Iran.
2- Department of Pathology, Shiraz University of Medical Sciences, Shiraz, Iran.
3- Persian Bayan Gene Research and Training Center, Dr. Faghihi’s Medical Genetic Center, Siraz, Iran. Shiraz Nephron-Urology Research Center, Shiraz University of Medical Sciences, Shiraz, Iran. ,yavarian@sums.ac.ir
2- Department of Pathology, Shiraz University of Medical Sciences, Shiraz, Iran.
3- Persian Bayan Gene Research and Training Center, Dr. Faghihi’s Medical Genetic Center, Siraz, Iran. Shiraz Nephron-Urology Research Center, Shiraz University of Medical Sciences, Shiraz, Iran. ,
Full-Text [PDF 382 kb]
(1120 Downloads)
| Abstract (HTML) (2709 Views)
In human, around 60 genes identified that is affected by the phenomenon of genetic imprinting (8, 9). Their expressions are critical during developmental fetal times. Many of them are regulatory genes involved in the growth and development of the fetus and are functionally hemizygous pattern. Therefore, alteration of effective copy number can cause developmental disorders.
Among these genes, the Insulin-like growth factor-2 (IGF-2) gene at the position 11p15.5 is paternally expressed in fetus (10). The position in chromosome includes an imprinting region that contains the IGF2 gene, which is consequently linked to the H19 gene. The IGF2/H19 expression is regulated by two imprinting control regions (ICR1 and ICR2). The ICR1 is a differentially methylated region at the upstream H19 promoter (11, 12). On the maternal allele, the CTCF protein, a zinc finger protein, binds to differentially methylated region (13, 14) and subsequently un-methylated maternal allele prevents IGF-2 interaction with an H19 enhancer at the downstream and inhibits IGF-2 expression. Methylation of this region on a paternal allele changes the expression of gene profile and favors IGF-2 expression (15). IGF-2 gene-encoded protein is involved in prenatal growth and is highly active during embryonic development. The activity of the IGF-2 gene depends on the copy that is inherited from the father (16). IGF-2 gene regulates the growth rate of the placenta and the embryo through angiogenesis (17-20), nutrient transfer (21), and inhibition of apoptosis (22), and its dysfunction can lead to miscarriage (23, 24). The IGF-2 gene has single nucleotide polymorphism in its different regions that affects its expression quantities and function. One of such known single nucleotide polymorphism is 820A/G variant (25). This variant in the father reduces gene expression and leads to abortion (26). The diagnosis of paternal 820 A/G-genotypes of the IGF-2 gene helps to predict the prospect of fetal loss due to gene imprinting.
This study was intended to inspect the frequency and potential effects of alleles in women with recurrent abortions to open a way to prevent this public burden.
About 1 cm of aborted fetal tissue was stored at -20ºC and used for DNA extraction. Samples were collected through questionnaire requiring information with spouse at the time of admission.
All samples were run along with positive and normal controls that were originally approved by Sanger sequencing method. Data were analyzed and genotyped using the HRM software.

2.4. Ethical consideration
The research was performed with the approval Ethics Committee of Islamic Azad University of Dehaghan Branch, Dehaghan, Iran (Code: IR.IAU.DEHAGHAN.REC.1397.001). Written informed consent was obtained from all candidates before taking samples.
2.5. Statistical analysis
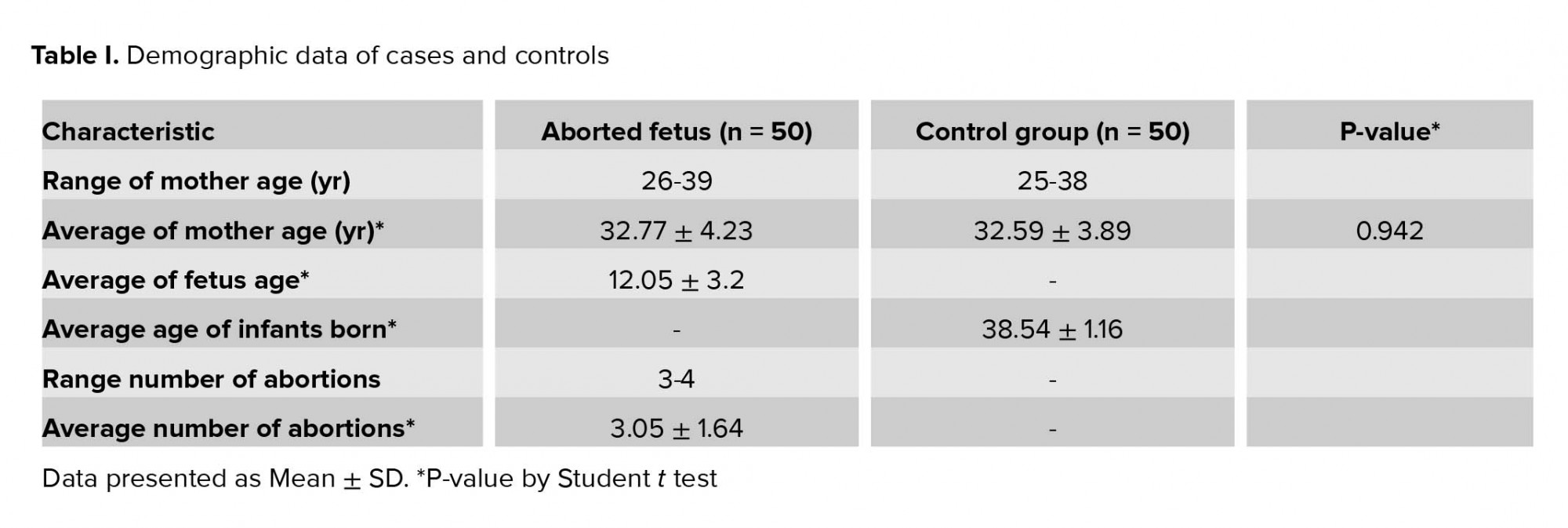
The statistical analysis was performed using the SPSS software (Statistical Package for the Social Sciences, version 22; SPSS, Chicago, IL, USA) and Chi-square (χ2) test by analysis of logistic regression; p < 0.05 was considered as the level of significance and A/G distribution was in agreement with the Hardy-Weinberg equilibrium (HWE). Odds ratio (OR) and p-value with 95% confidence interval (CI) were considered to estimate the risk RSA for 820A/G variant in the IGF-2 gene in the study group with RSA and control groups. Student’s t test was used to compare the mean age of the mothers between the two groups and Fisher's exact test for p-value.
3. Results
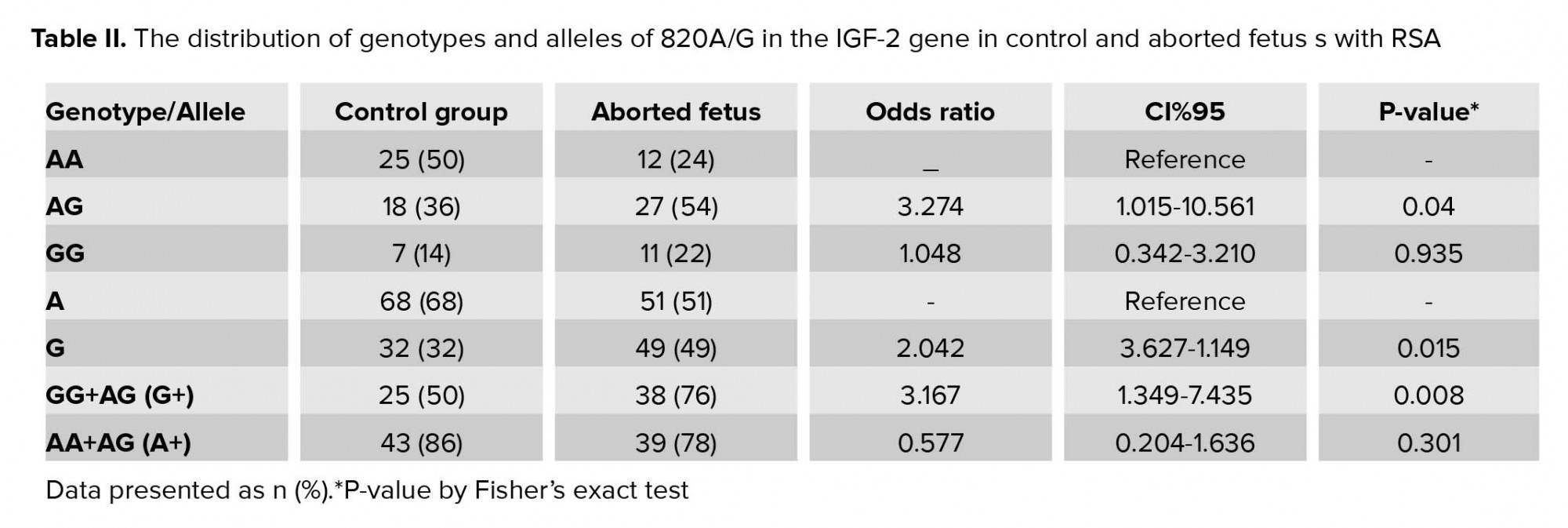
In present study, there was no significant difference between the mean age of mothers, the mothers of the control group with age 32.59± 3.89 yr compared to the mothers of aborted fetuses with age 32.77 ± 4.23 (p = 0.942) (Table I). The allelic and genotypic frequency of 820 A/G in the IGF-2 gene was studied in case and control groups. The ancestral allele A is considered as the reference. Based on the results of the statistical analysis revealed, the G allele is effective in recurrent abortions (p = 0.015). Considering the AA-genotype, as reference and analysis of other genotypes relative to this position obtained a statistically significant relationship (p = 0.04 for AG- genotype and p = 0.935 for GG-genotype). The AG genotype is effective in the incidence of recurrent miscarriage; the calculated OR showed that A/G genotype of the IGF-2 gene at 820 locus increased the incidence of spontaneous abortion more than three-fold. Considering the phenotypic status to the advantage of allele dominance in G, a significant relationship was obtained. The statistical analysis results are presented in Table II.
4. Discussion
The cause and mechanism underlying the traits and conditions that induce recurrent abortion is an important challenge for current genetics. Now, it is well-accepted that some human traits depend on the parent from whom the gene responsible for the trait is inherited. Obviously, the zygotes that were generated by maternally or paternally derived chromosomes could not survive to term. Development failure results from the entire chromosome complement inherited from only one parent. Comparison of gross morphologic complementarity of the phenotypes resulting from paternal sets of chromosomes versus maternal sets suggests that paternal genetic contribution is important for placental development, while maternal contribution is essential for proper embryo development (27).
Recently, frequent reports indications a connection between pregnancy complications such as restricted embryonic growth and RSA and the role of the IGF-2 gene. IGF-2 is known as a strong mitogen and is part of a cluster of imprinting genes on human chromosomes. Most imprinting genes are associated with embryo growth. The 820 A/G variation is a functional polymorphism and alter the primary sequence of encoded protein. This variation at the genomic level strongly affects the transcription level and alter IGF-2 expression status. Thus, mRNA transcription of the IGF-2 gene in the presence of this polymorphism will change quantatively. The G allele of the IGF-2 gene and plasma levels of IGF-2 protein as well as IGF-2 mRNA shows positive correlation. Therefore, heterozygote A/G at position 820 of this gene could results in loss of genomic imprinting manner in IGF-2 expression (25, 26, 28, 29).
The expression of the paternal allele is due to the changes in the epigenetic, including changes in the DNA structure such as DNA methylation. Imprinting genes or gene expression regulation is conducted via chromatin fiber change. The loss of the genetic imprinting (LOI) can result in a loss of the allele and imbalance (loss of function or increase in it) in a rate of the gene product and possibly also lead to the phenotypic outcome (30). Thus, the LOI leads to the loss of normal growth and development of the fetus. In order to clarify IGF-2 gene significance in the growth and development of pre-natal, Lighten and colleagues analyzed IGF genes and their receptors (IGF-1-IGF-2) expression in chorionic cells. Transcripts were evident that both genes and their receptors exist in human zygote cells before implantation. Although, according to the ligands, only transcripts of the IGF-2 ligand was detected. Based on their results, while before implantation, the parental allele function is blocked, genomic imprinting participated at the period of 8-cell embryo (31, 32). Therefore, it can be assumed that in RSA, the function of the genetic imprinting mechanism is not started, which results at the end of the growth of the organism (25).
Ostojic and co-worker have reviewed the genetic background of idiopathic RSA and the contribution of genetic changes to IGF-2 and H19 imprinting genes. This case-sectional study determined the relationship between IGF-2 820A/G and H19 HhaI gene variant RSA susceptibility. There was a significant difference in the frequency of IGF-2 820A/G in men with RSA compared to healthy men (p < 0.0001). There was no difference in the distribution of this genotypic women's groups. The presence of IGF-2 the 820 variant among the husbands of these women in RSA couples can affect the expression of IGF-2 in the placenta and embryo and represents a risk factor for RSA sensitivity (26). In addition, a study of 107 placental tissue samples from Ukraine reported that aborted embryos at 10-15 wk of pregnancy, carrier fetus with 820 A/G genotype, has seven times more chance to develop RSA as compared to the AA genotype (25). This study is also in line with the findings of Koukoura and colleagues from Greece who studied 31 placentas with fetal growth restriction (FGR) and found decline in IGF2 mRNA levels and LOI among the abnormal placentas. The epigenetic mechanism that regulates the genetic imprinting of the IGF-2 gene leads to FGR and induce significantly the reduction at the level of IGF-2 mRNA (15).
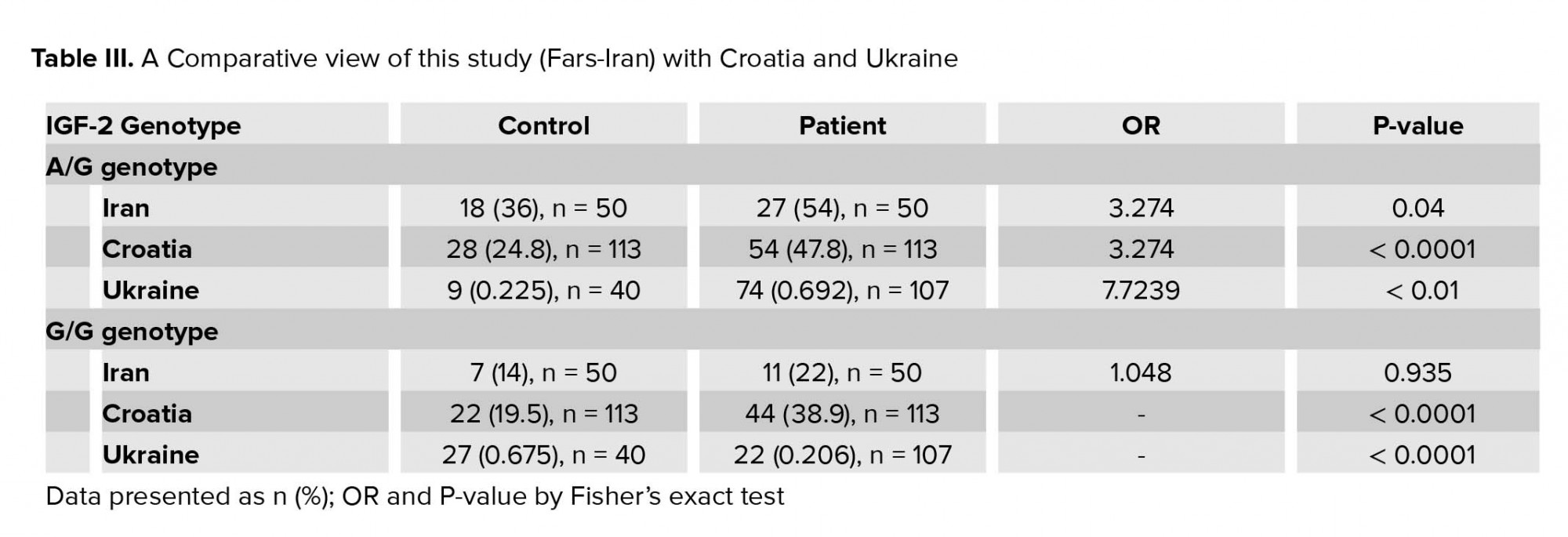
Although our study shows that the chance of RSA due to paternal G allele is increased about three times, this finding is still meaningfully lower than expected in comparison to the other study (Table III). The high rate of consanguinity marriage may introduce other possible genetic factors contributing to the development of RSA.
5. Conclusion
This study indicates that carriers of GG or A/G genotype are three times more likely to have recurrent abortion in our population.
Acknowledgments
The authors are thankful to all their colleagues who collaborated in this study. This research was supported by Islamic Azad University, Arsanjan, Iran.
Conflict of interest
There is no conflict of interest in this study.
Full-Text: (524 Views)
- Introduction
In human, around 60 genes identified that is affected by the phenomenon of genetic imprinting (8, 9). Their expressions are critical during developmental fetal times. Many of them are regulatory genes involved in the growth and development of the fetus and are functionally hemizygous pattern. Therefore, alteration of effective copy number can cause developmental disorders.
Among these genes, the Insulin-like growth factor-2 (IGF-2) gene at the position 11p15.5 is paternally expressed in fetus (10). The position in chromosome includes an imprinting region that contains the IGF2 gene, which is consequently linked to the H19 gene. The IGF2/H19 expression is regulated by two imprinting control regions (ICR1 and ICR2). The ICR1 is a differentially methylated region at the upstream H19 promoter (11, 12). On the maternal allele, the CTCF protein, a zinc finger protein, binds to differentially methylated region (13, 14) and subsequently un-methylated maternal allele prevents IGF-2 interaction with an H19 enhancer at the downstream and inhibits IGF-2 expression. Methylation of this region on a paternal allele changes the expression of gene profile and favors IGF-2 expression (15). IGF-2 gene-encoded protein is involved in prenatal growth and is highly active during embryonic development. The activity of the IGF-2 gene depends on the copy that is inherited from the father (16). IGF-2 gene regulates the growth rate of the placenta and the embryo through angiogenesis (17-20), nutrient transfer (21), and inhibition of apoptosis (22), and its dysfunction can lead to miscarriage (23, 24). The IGF-2 gene has single nucleotide polymorphism in its different regions that affects its expression quantities and function. One of such known single nucleotide polymorphism is 820A/G variant (25). This variant in the father reduces gene expression and leads to abortion (26). The diagnosis of paternal 820 A/G-genotypes of the IGF-2 gene helps to predict the prospect of fetal loss due to gene imprinting.
This study was intended to inspect the frequency and potential effects of alleles in women with recurrent abortions to open a way to prevent this public burden.
- Materials and Methods
- 1. Samples
About 1 cm of aborted fetal tissue was stored at -20ºC and used for DNA extraction. Samples were collected through questionnaire requiring information with spouse at the time of admission.
- 2. Genetic analysis
All samples were run along with positive and normal controls that were originally approved by Sanger sequencing method. Data were analyzed and genotyped using the HRM software.
- 3. Real-time PCR reactions and melting analysis


2.4. Ethical consideration
The research was performed with the approval Ethics Committee of Islamic Azad University of Dehaghan Branch, Dehaghan, Iran (Code: IR.IAU.DEHAGHAN.REC.1397.001). Written informed consent was obtained from all candidates before taking samples.
2.5. Statistical analysis
The statistical analysis was performed using the SPSS software (Statistical Package for the Social Sciences, version 22; SPSS, Chicago, IL, USA) and Chi-square (χ2) test by analysis of logistic regression; p < 0.05 was considered as the level of significance and A/G distribution was in agreement with the Hardy-Weinberg equilibrium (HWE). Odds ratio (OR) and p-value with 95% confidence interval (CI) were considered to estimate the risk RSA for 820A/G variant in the IGF-2 gene in the study group with RSA and control groups. Student’s t test was used to compare the mean age of the mothers between the two groups and Fisher's exact test for p-value.
3. Results
In present study, there was no significant difference between the mean age of mothers, the mothers of the control group with age 32.59± 3.89 yr compared to the mothers of aborted fetuses with age 32.77 ± 4.23 (p = 0.942) (Table I). The allelic and genotypic frequency of 820 A/G in the IGF-2 gene was studied in case and control groups. The ancestral allele A is considered as the reference. Based on the results of the statistical analysis revealed, the G allele is effective in recurrent abortions (p = 0.015). Considering the AA-genotype, as reference and analysis of other genotypes relative to this position obtained a statistically significant relationship (p = 0.04 for AG- genotype and p = 0.935 for GG-genotype). The AG genotype is effective in the incidence of recurrent miscarriage; the calculated OR showed that A/G genotype of the IGF-2 gene at 820 locus increased the incidence of spontaneous abortion more than three-fold. Considering the phenotypic status to the advantage of allele dominance in G, a significant relationship was obtained. The statistical analysis results are presented in Table II.

4. Discussion
The cause and mechanism underlying the traits and conditions that induce recurrent abortion is an important challenge for current genetics. Now, it is well-accepted that some human traits depend on the parent from whom the gene responsible for the trait is inherited. Obviously, the zygotes that were generated by maternally or paternally derived chromosomes could not survive to term. Development failure results from the entire chromosome complement inherited from only one parent. Comparison of gross morphologic complementarity of the phenotypes resulting from paternal sets of chromosomes versus maternal sets suggests that paternal genetic contribution is important for placental development, while maternal contribution is essential for proper embryo development (27).
Recently, frequent reports indications a connection between pregnancy complications such as restricted embryonic growth and RSA and the role of the IGF-2 gene. IGF-2 is known as a strong mitogen and is part of a cluster of imprinting genes on human chromosomes. Most imprinting genes are associated with embryo growth. The 820 A/G variation is a functional polymorphism and alter the primary sequence of encoded protein. This variation at the genomic level strongly affects the transcription level and alter IGF-2 expression status. Thus, mRNA transcription of the IGF-2 gene in the presence of this polymorphism will change quantatively. The G allele of the IGF-2 gene and plasma levels of IGF-2 protein as well as IGF-2 mRNA shows positive correlation. Therefore, heterozygote A/G at position 820 of this gene could results in loss of genomic imprinting manner in IGF-2 expression (25, 26, 28, 29).
The expression of the paternal allele is due to the changes in the epigenetic, including changes in the DNA structure such as DNA methylation. Imprinting genes or gene expression regulation is conducted via chromatin fiber change. The loss of the genetic imprinting (LOI) can result in a loss of the allele and imbalance (loss of function or increase in it) in a rate of the gene product and possibly also lead to the phenotypic outcome (30). Thus, the LOI leads to the loss of normal growth and development of the fetus. In order to clarify IGF-2 gene significance in the growth and development of pre-natal, Lighten and colleagues analyzed IGF genes and their receptors (IGF-1-IGF-2) expression in chorionic cells. Transcripts were evident that both genes and their receptors exist in human zygote cells before implantation. Although, according to the ligands, only transcripts of the IGF-2 ligand was detected. Based on their results, while before implantation, the parental allele function is blocked, genomic imprinting participated at the period of 8-cell embryo (31, 32). Therefore, it can be assumed that in RSA, the function of the genetic imprinting mechanism is not started, which results at the end of the growth of the organism (25).
Ostojic and co-worker have reviewed the genetic background of idiopathic RSA and the contribution of genetic changes to IGF-2 and H19 imprinting genes. This case-sectional study determined the relationship between IGF-2 820A/G and H19 HhaI gene variant RSA susceptibility. There was a significant difference in the frequency of IGF-2 820A/G in men with RSA compared to healthy men (p < 0.0001). There was no difference in the distribution of this genotypic women's groups. The presence of IGF-2 the 820 variant among the husbands of these women in RSA couples can affect the expression of IGF-2 in the placenta and embryo and represents a risk factor for RSA sensitivity (26). In addition, a study of 107 placental tissue samples from Ukraine reported that aborted embryos at 10-15 wk of pregnancy, carrier fetus with 820 A/G genotype, has seven times more chance to develop RSA as compared to the AA genotype (25). This study is also in line with the findings of Koukoura and colleagues from Greece who studied 31 placentas with fetal growth restriction (FGR) and found decline in IGF2 mRNA levels and LOI among the abnormal placentas. The epigenetic mechanism that regulates the genetic imprinting of the IGF-2 gene leads to FGR and induce significantly the reduction at the level of IGF-2 mRNA (15).
Although our study shows that the chance of RSA due to paternal G allele is increased about three times, this finding is still meaningfully lower than expected in comparison to the other study (Table III). The high rate of consanguinity marriage may introduce other possible genetic factors contributing to the development of RSA.

5. Conclusion
This study indicates that carriers of GG or A/G genotype are three times more likely to have recurrent abortion in our population.
Acknowledgments
The authors are thankful to all their colleagues who collaborated in this study. This research was supported by Islamic Azad University, Arsanjan, Iran.
Conflict of interest
There is no conflict of interest in this study.
Type of Study: Original Article |
Subject:
Fertility & Infertility
References
1. Matthiesen L, Kalkunte S, Sharma S. Multiple pregnancy failures: an immunological paradigm. Am J Reprod Immunol 2012; 67: 334-340. [DOI:10.1111/j.1600-0897.2012.01121.x] [PMID]
2. McNamee K, Dawood F, Farquharson R. Recurrent miscarriage and thrombophilia: An update. Curr Opin Obstet Gynecol 2012; 24: 229-234. [DOI:10.1097/GCO.0b013e32835585dc] [PMID]
3. Roland L, Gagne A, Belanger MC, Boutet M, Julien P, Bilodeau JF. Plasma interleukin-18 (IL- 18) levels are correlated with antioxidant vitamin coenzyme Q (10) in preeclampsia. Acta Obstet Gynecol Scand 2010; 89: 360-366. [DOI:10.3109/00016340903576020] [PMID]
4. Huang HY. The cytokine network during embryo implantation. Chang Gung Med J 2006; 29: 25-36.
5. Frequently asked questions: pregnancy. repeated miscarriages. The American College of Obstetricians and Gynecologists. Available at: URL: https://m.acog.org/Patients/FAQs/Repeated-Miscarriages.
6. Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 2002; 62: 6442-6446.
7. Gurrieri F, Accadia M. Genetic imprinting: the paradigm of prader-willi and angelman syndromes. Endocr Dev 2009; 14: 20-28. [DOI:10.1159/000207473] [PMID]
8. Moore GE, Oakey R. The role of imprinted genes in humans. Genome Biol 2011; 12: 106. [DOI:10.1186/gb-2011-12-3-106] [PMID] [PMCID]
9. Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet 2003; 12: 535-549. [DOI:10.1093/hmg/ddg034] [PMID]
10. Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol 2002; 192: 245-258. [DOI:10.1002/jcp.10129] [PMID]
11. Horike SI, Ferreira JCP, Meguro-Horike M, Choufani S, Smith AC, Shuman C, et al. Screening of DNA methylation at the H19 promoter or the distal region of its ICR1 ensures efficient detection of chromosome 11p15 epimutations in Russell-Silver syndrome. Am J Med Genet A 2009; 149: 2415-2423. [DOI:10.1002/ajmg.a.33065] [PMID]
12. Netchine I, Rossignol S, Dufourg MN, Azzi S, Rousseau A, Perin L, et al. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab 2007; 92: 3148-3154. [DOI:10.1210/jc.2007-0354] [PMID]
13. Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 1996; 16: 2802-2813. [DOI:10.1128/MCB.16.6.2802] [PMID] [PMCID]
14. Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohes in to chromatin. Proc Natl Acad Sci USA 2008; 105: 8309-8314. [DOI:10.1073/pnas.0801273105] [PMID] [PMCID]
15. Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, Jones A, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med 2011; 28: 481-487.
16. IGF2 gene. Genetics Home Reference. Available at: URL:https://ghr.nlm.nih.gov/gene/IGF2.
17. McKinnon T, Chakraborty C, Gleeson LM, Chidiac P, Lala PK. Stimulation of human extra villous trophoblast migration by IGF-II is mediated by IGF type 2 receptor involving inhibitory G protein (s) and phosphorylation of MAPK. J Clin Endocrinol Metab 2001; 86: 3665-3674. [DOI:10.1210/jcem.86.8.7711] [PMID]
18. Lee OH, Bae SK, Bae MH, Lee YM, Moon EJ, Cha HJ, et al. Identification of angiogenic properties of insulin-like growth factor II in in vitro angiogenesis models. Br J Cancer 2000; 82: 385-391. [DOI:10.1054/bjoc.1999.0931] [PMID] [PMCID]
19. Hills FA, Elder MG, Chard T, Sullivan MHF. Regulation of human villous trophoblast by insulin-like growth factors and insulin-like growth factor-binding protein-1. J Endocrinol 2004; 183: 487-496. [DOI:10.1677/joe.1.05867] [PMID]
20. Herr F, Liang OD, Herrero J, Lang U, Preissner KT, Han VK, et al. Possible angiogenic roles of insulin-like growth factor II and its receptors in uterine vascular adaptation to pregnancy. J Clin Endocrinol Metab 2003; 88: 4811-4817. [DOI:10.1210/jc.2003-030243] [PMID]
21. Reik W, Constancia M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, et al. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol 2003; 547: 35-44. [DOI:10.1113/jphysiol.2002.033274] [PMID] [PMCID]
22. Stewart CE, Rotwein P. Insulin-like growth factor-II is an autocrine survival factor for differentiating myoblasts. J Biol Chem 1996; 271: 11330-11338. [DOI:10.1074/jbc.271.19.11330] [PMID]
23. Soejima H, Yun K. Allele-specific-polymerase chain reaction: a novel method for investigation of the imprinted insulin-like growth factor II gene. Lab Invest 1998; 78: 641-642.
24. Stray-Pedersen B, Stray-Pedersen S. Etiologic factors and subsequent reproductive performance in 195 couples with a prior history of habitual abortion. Am J Obstet Gynecol 1984; 148: 140-146. [DOI:10.1016/S0002-9378(84)80164-7]
25. Zastavna D, Makukh H, Tretjak B, Bilevych O, Tyrka M. Loss of Imprinting of IGF2 Gene in the Chorionic Tissues of Spontaneously 21-Eliminated Human Embryos. Genet Epigenet 2013; 5: 17-22. [DOI:10.4137/GEG.S11460] [PMID] [PMCID]
26. Ostojic S, Pereza N, Volk M, Kapovic M, Peterlin B. Genetic predisposition to idiopathic recurrent spontaneous abortion: contribution of genetic variations in IGF-2 and H19 imprinted genes. Am J Reprod Immunol 2008; 60: 111-117. [DOI:10.1111/j.1600-0897.2008.00601.x] [PMID]
27. Solter D, Aronson J, Gilbert SF, McGrath J. Nuclear transfer in mouse embryos: activation of the embryonic genome. Cold Spring Harb Symp Quant Biol 1985; 50: 45-50. [DOI:10.1101/SQB.1985.050.01.008] [PMID]
28. Rodriguez S, Gaunt TR, O Dell SD, Chen XH, Gu D, Hawe E, et al. Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardio vascular risk traits. Hum Mol Genet 2004; 13: 715-725. [DOI:10.1093/hmg/ddh070] [PMID]
29. Sandhu MS, Gibson JM, Heald AH, Dunger DB, Wareham NJ. Low circulating IGF-II concentrations predict weight gain and obesity in humans. Diabetes 2003; 52: 1403-1408. [DOI:10.2337/diabetes.52.6.1403] [PMID]
30. Chen J, Fang Q, Chen B, Zhou Y, Luo Y. Study on the imprinting status of insulin-like growth factor II (IGF-II) gene in villus during 6-10 gestational weeks. Obstet Gynecol Int 2010; 2010: 965905-965908. [DOI:10.1155/2010/965905] [PMID] [PMCID]
31. Lighten AD, Hardy K, Winston RM, Moore GE. IGF2 is parentally imprinted in human preimplantation embryos. Nat Genet 1997; 15: 122-123. [DOI:10.1038/ng0297-122] [PMID]
32. Lighten AD, Hardy K, Winston RM, Moore GE. Expression of mRNA for the insulin-like growth factors and their receptors in human preimplantation embryos. Mol Reprod Dev 1997; 47: 134-139.
https://doi.org/10.1002/(SICI)1098-2795(199706)47:2<134::AID-MRD2>3.0.CO;2-N [DOI:10.1002/(SICI)1098-2795(199706)47:23.0.CO;2-N]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





