Wed, Apr 24, 2024
[Archive]
Volume 20, Issue 7 (July 2022)
IJRM 2022, 20(7): 561-568 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Barzanouni S, Moramezi F, Zargar M, Galehdari H, Hemadi M. A prospective study to evaluate the gender prediction of blastocysts by using cell-free DNA within a culture medium. IJRM 2022; 20 (7) :561-568
URL: http://ijrm.ir/article-1-2120-en.html
URL: http://ijrm.ir/article-1-2120-en.html
1- Department of Obstetrics and Gynecology, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. , drbarzanonis@gmail.com
2- Department of Obstetrics and Gynecology, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
3- Department of Obstetrics and Gynecology, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
4- Department of Genetics, Faculty of Sciences, Shahid Chamran University of Ahvaz, Ahvaz, Iran.
5- Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
2- Department of Obstetrics and Gynecology, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
3- Department of Obstetrics and Gynecology, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
4- Department of Genetics, Faculty of Sciences, Shahid Chamran University of Ahvaz, Ahvaz, Iran.
5- Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Keywords: Preimplantation diagnosis, Embryo implantation, Culture media, Blastocyst, Polymerase chain reaction.
Full-Text [PDF 409 kb]
(596 Downloads)
| Abstract (HTML) (842 Views)
Full-Text: (134 Views)
- Introduction
One of the essential functions of using in vitro fertilization (IVF) techniques is to raise the possibility of having a healthy child in couples who are in danger of having a child with a genetic disease (1, 2). To accomplish this, before embryo implantation in the uterus, the genetic health of the embryo produced in laboratory conditions is examined using preimplantation genetic diagnosis (PGD). They are then transferred to the uterus after assuring genetic health (1-4).
The common method of PGD implementation includes controlled ovarian stimulation, egg acquisition, intracytoplasmic sperm injection in egg, keeping the embryos in culture medium, detachment of one cell from an embryo on day 3 or a few cells on day 5 for genetic diagnosis, performing the necessary tests to diagnose, and finally select an embryo without disease and transfer to the uterus (4-6). This method unlike other diagnostic methods does not cause complications that may lead to termination of pregnancy (3). However, this method of PGD due to cell separation from the embryo is a kind of invasive method then it can cause abortion after implantation (7, 8). Recently, 2 noninvasive methods for implementing PGD for achieving embryo genetic material have been introduced, including PGD using blastocysts fluid and PGD using embryo culture medium (7-11). In both approaches, the required embryonic DNA is achieved from matters discarded after implantation in the uterus. The main drawback of both approaches is access to a small amount of genetic material in the embryo, making the performance of genetic studies difficult (7, 9). According to a survey, it was estimated that 80 ± 70 and 99 ± 113 pg of DNA were present in the medium on day 3 and day 5/6, respectively. The study showed that with this amount of DNA, the TBC1 domain family member 3 gene, which is located on chromosome 17, could be identified, but not enough to accurately identify the testis-specific protein Y-Linked 1 gene, which is located on the Y chromosome (12). It was reported that combining the amount of DNA obtained from both methods is sufficient for whole genome amplification and accurate aneuploidy screening (9).
Due to medical reasons, gender selection is defined as preventing childbirth with a severe genetic disorder (13, 14). PGD was initially introduced for selecting female embryos concerning chromosome X-related disorders (5, 14, 15). Although, currently, some families, due to several issues such as having multiple children of the same sex, wanting to have a child of different gender, request IVF implementation and pre-implantation genetic diagnosis (16).
This prospective clinical trial study attempts to determine the gender of embryos formed in the laboratory on day 5, using embryonic DNA extraction from the culture medium, and evaluation of the sex-determining region Y (SRY) and fragile X mental retardation (FMR1) genes for diagnosis of Y and X chromosome, respectively using the polymerase chain reaction (PCR) technique (17-19). Then, after the diagnosis of the gender of implanted embryos in the 4th month of pregnancy by ultrasound, the accuracy of this method was evaluated.
The common method of PGD implementation includes controlled ovarian stimulation, egg acquisition, intracytoplasmic sperm injection in egg, keeping the embryos in culture medium, detachment of one cell from an embryo on day 3 or a few cells on day 5 for genetic diagnosis, performing the necessary tests to diagnose, and finally select an embryo without disease and transfer to the uterus (4-6). This method unlike other diagnostic methods does not cause complications that may lead to termination of pregnancy (3). However, this method of PGD due to cell separation from the embryo is a kind of invasive method then it can cause abortion after implantation (7, 8). Recently, 2 noninvasive methods for implementing PGD for achieving embryo genetic material have been introduced, including PGD using blastocysts fluid and PGD using embryo culture medium (7-11). In both approaches, the required embryonic DNA is achieved from matters discarded after implantation in the uterus. The main drawback of both approaches is access to a small amount of genetic material in the embryo, making the performance of genetic studies difficult (7, 9). According to a survey, it was estimated that 80 ± 70 and 99 ± 113 pg of DNA were present in the medium on day 3 and day 5/6, respectively. The study showed that with this amount of DNA, the TBC1 domain family member 3 gene, which is located on chromosome 17, could be identified, but not enough to accurately identify the testis-specific protein Y-Linked 1 gene, which is located on the Y chromosome (12). It was reported that combining the amount of DNA obtained from both methods is sufficient for whole genome amplification and accurate aneuploidy screening (9).
Due to medical reasons, gender selection is defined as preventing childbirth with a severe genetic disorder (13, 14). PGD was initially introduced for selecting female embryos concerning chromosome X-related disorders (5, 14, 15). Although, currently, some families, due to several issues such as having multiple children of the same sex, wanting to have a child of different gender, request IVF implementation and pre-implantation genetic diagnosis (16).
This prospective clinical trial study attempts to determine the gender of embryos formed in the laboratory on day 5, using embryonic DNA extraction from the culture medium, and evaluation of the sex-determining region Y (SRY) and fragile X mental retardation (FMR1) genes for diagnosis of Y and X chromosome, respectively using the polymerase chain reaction (PCR) technique (17-19). Then, after the diagnosis of the gender of implanted embryos in the 4th month of pregnancy by ultrasound, the accuracy of this method was evaluated.
- Material and Methods
- Sample collection and preparation
This study was performed between December 2018 and March 2020 in Imam Khomeyni hospital, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
According to the Cochran test, the minimum number of embryos required was 23. Due to the possibility of losing samples during the study, 30 samples were examined in the study.
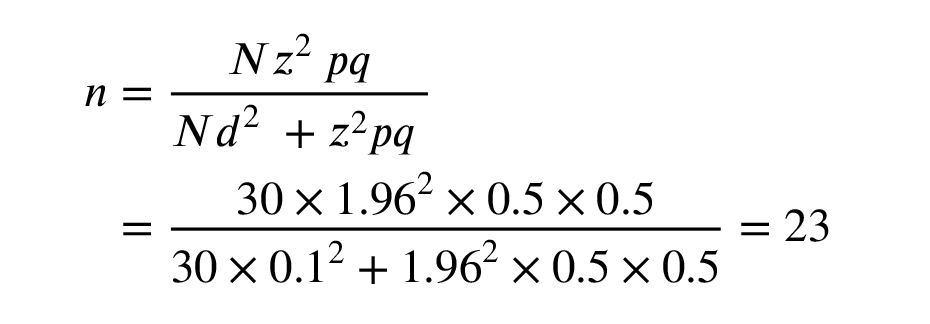
The inclusion criteria were couples referred to the fertility clinic of Imam Khomeyni hospital for infertility and seeking assisted reproductive technology and IVF assistance using PGD for determining the gender of embryos, with female partners < 35 yr old.
The exclusion criteria were couples with cancelled cooperation, unsuccessful IVF, and no pregnancy or abortion before the 4th month of pregnancy. If the IVF was unsuccessful and the pregnancy was terminated before birth, a new couple would be substituted.
Couples underwent transvaginal oocyte retrieval and fertilization using intracytoplasmic sperm injection. Following the intracytoplasmic sperm injection, fertilized oocytes were cultured in G-TL™ media (Vitrolife, Sweden) at 37ºC with 6% CO2 and 5% O2. A total of 30 preimplantation embryos (calculated based on Cochran test) at day 5 were examined. After the embryo was removed, 20 μl of media were recovered from each day 5 embryo culture and transferred to DNA-free/DNase-free tubes under sterile conditions. DNA extraction and amplification were performed using the REPLI-g Single Cell Kit (QIAGEN, Netherlands) following the manufacture’s protocol. The amplification product was quantified with nanodrop. Finally, the samples were stocked at 4ºC until PCR.
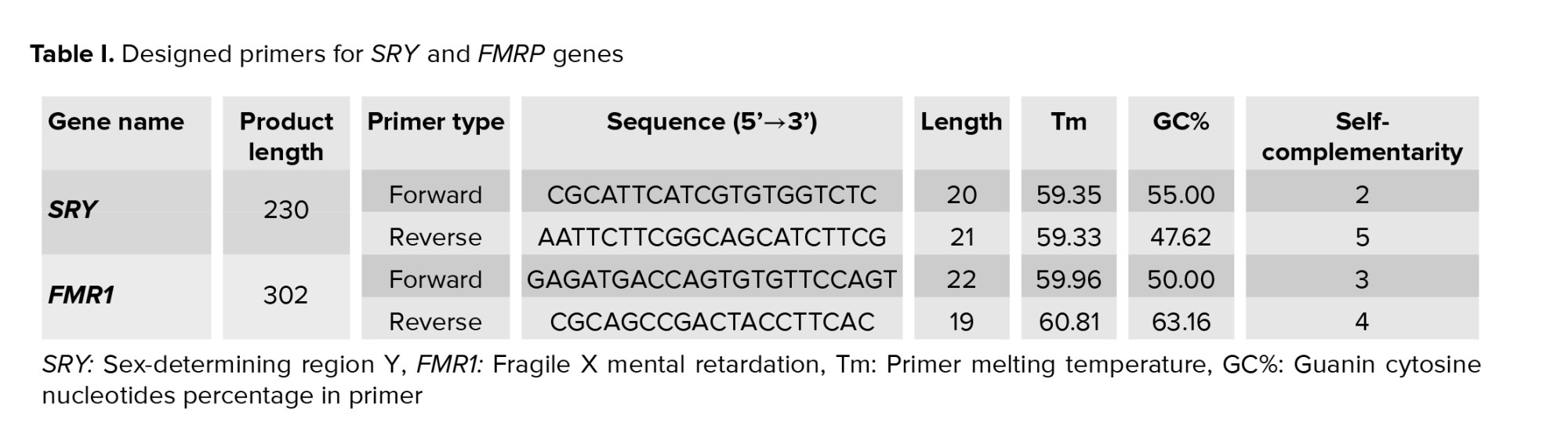
The SRY and FMR1 genes were successfully amplified in day 5 culture medium and detected with bands at 230 bp and 301 bp, respectively. Positive controls (male and female DNA) were included in the PCR reactions. If PCR products were observed at both 230 bp and 301 bp, this result indicated that the DNA originated from a male sample. However, if samples originated from a female-only, a single PCR product at 301 bp was observed (Figure 1).
Analysis of PCR products showed that out of 30 embryos studied, 7 were male, and 23 were female. However, an ultrasound revealed that 13 fetuses were male and 17 were female. Thus, in 6 male embryos, the Y chromosome was not detected by amplified SRY gene, and sex determination was incorrect. The calculation of the sensitivity, specificity, and diagnostic accuracy of this method was done based on the ‘2 by 2 table’ and the results are as follows (Table III):
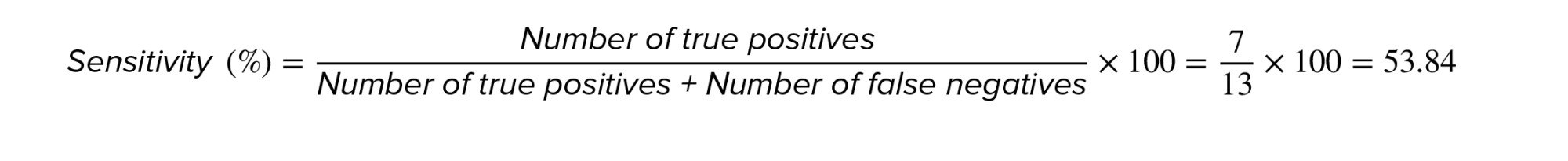
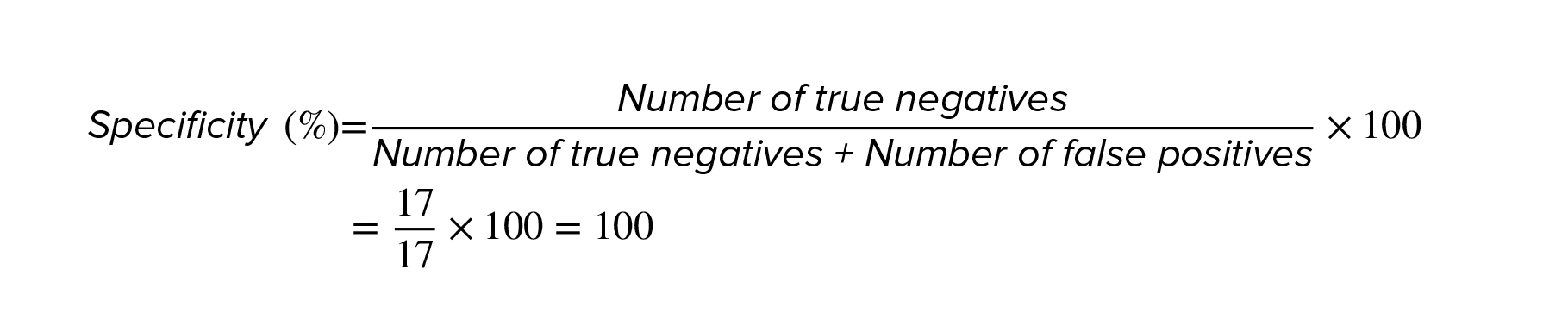

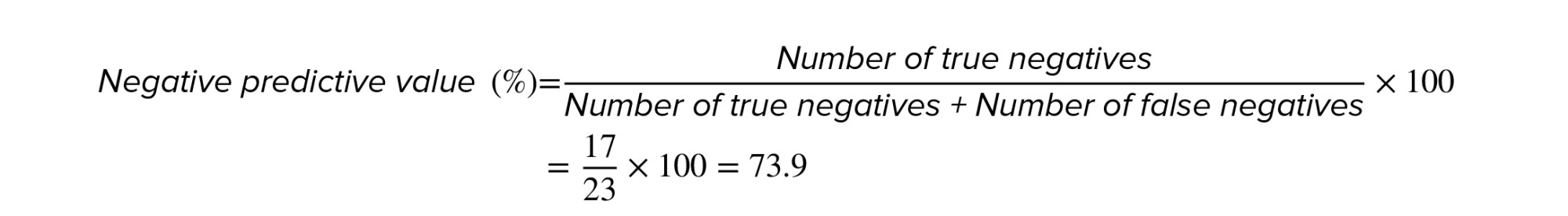


As a result, it can be said that although the gender of all 7 cases diagnosed by ultrasound was the same as found in the culture medium, the gender of the diagnosed girls had an accuracy of 74%. So, in case, a male gender is desired, we can trust the method to determine the gender of the embryo. However, if the preferred gender is female, it should be considered that 26% of the cases diagnosed as females using this method are probably males.
One of the factors that can justify this result is paying attention to the size and dimensions of the 2 chromosomes X and Y. The Y chromosome is one of the smallest human chromosomes with approximately 57.23 nucleotides, and the SRY gene located at the Yp11.2 locus comprises only 828 nucleotides. In contrast, the X chromosome is about 156.05 Mbp long (2.72 times the Y chromosome), and the FMR1 gene, located at locus Xq27.3, has 46,135 nucleotides (about 55.71 times larger than the SRY gene).
A previous study used multiplex PCR for the detection of SRY (as Y chromosome gene) and ALT1 genes (as control). This study was performed on DNA extracted from blood cells. Its purpose was to evaluate the efficiency of multiplex PCR technique and the sensitivity of SRY gene to detect the male gender (18).
Another study reported that the use of cell-free DNA in the culture medium may be a viable alternative to the use of embryonic cell DNA for the diagnosis of a-thalassemia-SEA (20). The study evaluated the amount of cell-free DNA in the culture medium from the culture on day 3 and day 5, and reported that it was better to use the day 5 medium due to the higher amount of cell-free DNA.
Furthermore, Li et al. showed that cell-free DNA in culture medium or fetal blastocyst fluid could be used to assess genetic and chromosomal defects. However, the DNA obtained from these methods is small, and it may be necessary to use both ways together (7).
According to Yang et al., the amount of cell-free DNA in day 3 embryonic culture medium was insufficient to detect the Y chromosome by the SRY gene, and it was recommended to use the day 5 embryonic culture medium (8).
Given the huge difference in the dimensions of these 2 chromosomes, it is quite clear that the extraction of the Y chromosome and the study of its genes is more difficult than the extraction and study of the genes of the X chromosome, particularly in the extraction of embryo's genetic material method from the culture medium where the amount of DNA obtained for examination is also little.
Our finding is consistent with the study that showed that the amount of DNA obtained from culture media was not sufficient for detecting accurately the testis-specific protein Y-Linked 1 gene, which is located on the Y chromosome but was enough for detecting the TBC1 domain family member 3 gene, which is located on chromosome 17 (12).
Acknowledgements
This work was supported by grants from the Infertility Center, Imam Khomeini hospital, Ahvaz Jundishapur University of Medical Sciences, Khuzestan, Iran.
Conflict of Interest
The authors declare that they have no conflicts of interest.
According to the Cochran test, the minimum number of embryos required was 23. Due to the possibility of losing samples during the study, 30 samples were examined in the study.
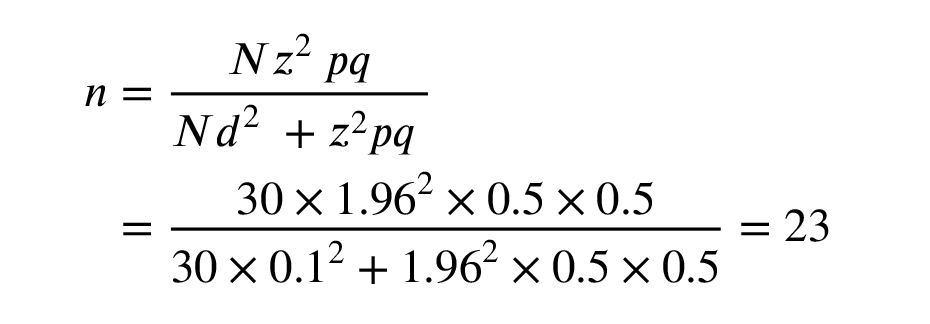
The inclusion criteria were couples referred to the fertility clinic of Imam Khomeyni hospital for infertility and seeking assisted reproductive technology and IVF assistance using PGD for determining the gender of embryos, with female partners < 35 yr old.
The exclusion criteria were couples with cancelled cooperation, unsuccessful IVF, and no pregnancy or abortion before the 4th month of pregnancy. If the IVF was unsuccessful and the pregnancy was terminated before birth, a new couple would be substituted.
Couples underwent transvaginal oocyte retrieval and fertilization using intracytoplasmic sperm injection. Following the intracytoplasmic sperm injection, fertilized oocytes were cultured in G-TL™ media (Vitrolife, Sweden) at 37ºC with 6% CO2 and 5% O2. A total of 30 preimplantation embryos (calculated based on Cochran test) at day 5 were examined. After the embryo was removed, 20 μl of media were recovered from each day 5 embryo culture and transferred to DNA-free/DNase-free tubes under sterile conditions. DNA extraction and amplification were performed using the REPLI-g Single Cell Kit (QIAGEN, Netherlands) following the manufacture’s protocol. The amplification product was quantified with nanodrop. Finally, the samples were stocked at 4ºC until PCR.
-
- PCR assay
- SRY+ / FMR1+, blastocyst has both X and Y chromosomes, blastocyst gender should be male.
- SRY- / FMR1+, blastocyst has only an X chromosome, blastocyst gender should be female.
- SRY+ / FMR1-, blastocyst has only a Y chromosome, maybe blastocyst has aneuploidy, the test should be repeated.
- SRY- / FMR1-, blastocyst have neither X nor Y chromosomes, the test should be repeated.


-
- Ethical considerations
- Results
The SRY and FMR1 genes were successfully amplified in day 5 culture medium and detected with bands at 230 bp and 301 bp, respectively. Positive controls (male and female DNA) were included in the PCR reactions. If PCR products were observed at both 230 bp and 301 bp, this result indicated that the DNA originated from a male sample. However, if samples originated from a female-only, a single PCR product at 301 bp was observed (Figure 1).
Analysis of PCR products showed that out of 30 embryos studied, 7 were male, and 23 were female. However, an ultrasound revealed that 13 fetuses were male and 17 were female. Thus, in 6 male embryos, the Y chromosome was not detected by amplified SRY gene, and sex determination was incorrect. The calculation of the sensitivity, specificity, and diagnostic accuracy of this method was done based on the ‘2 by 2 table’ and the results are as follows (Table III):
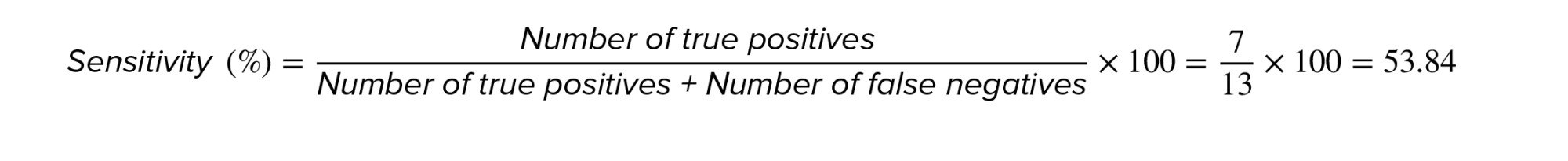
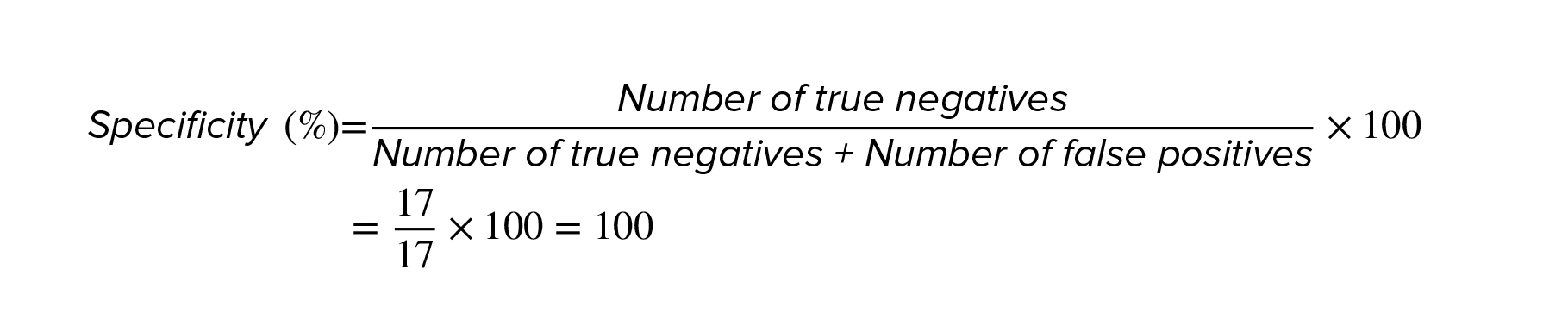

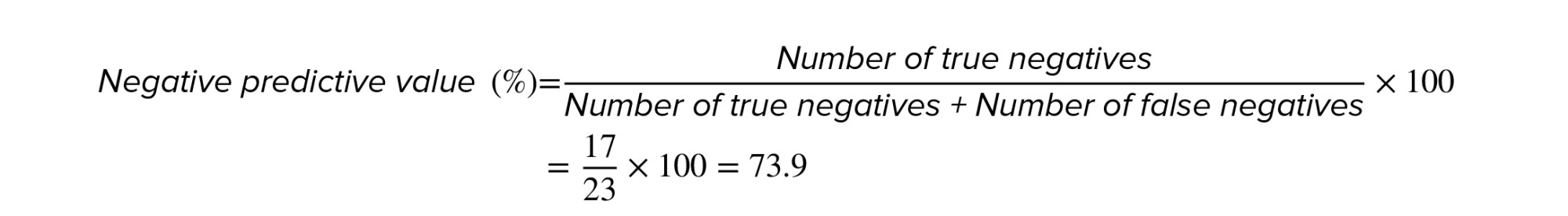


- Discussion
As a result, it can be said that although the gender of all 7 cases diagnosed by ultrasound was the same as found in the culture medium, the gender of the diagnosed girls had an accuracy of 74%. So, in case, a male gender is desired, we can trust the method to determine the gender of the embryo. However, if the preferred gender is female, it should be considered that 26% of the cases diagnosed as females using this method are probably males.
One of the factors that can justify this result is paying attention to the size and dimensions of the 2 chromosomes X and Y. The Y chromosome is one of the smallest human chromosomes with approximately 57.23 nucleotides, and the SRY gene located at the Yp11.2 locus comprises only 828 nucleotides. In contrast, the X chromosome is about 156.05 Mbp long (2.72 times the Y chromosome), and the FMR1 gene, located at locus Xq27.3, has 46,135 nucleotides (about 55.71 times larger than the SRY gene).
A previous study used multiplex PCR for the detection of SRY (as Y chromosome gene) and ALT1 genes (as control). This study was performed on DNA extracted from blood cells. Its purpose was to evaluate the efficiency of multiplex PCR technique and the sensitivity of SRY gene to detect the male gender (18).
Another study reported that the use of cell-free DNA in the culture medium may be a viable alternative to the use of embryonic cell DNA for the diagnosis of a-thalassemia-SEA (20). The study evaluated the amount of cell-free DNA in the culture medium from the culture on day 3 and day 5, and reported that it was better to use the day 5 medium due to the higher amount of cell-free DNA.
Furthermore, Li et al. showed that cell-free DNA in culture medium or fetal blastocyst fluid could be used to assess genetic and chromosomal defects. However, the DNA obtained from these methods is small, and it may be necessary to use both ways together (7).
According to Yang et al., the amount of cell-free DNA in day 3 embryonic culture medium was insufficient to detect the Y chromosome by the SRY gene, and it was recommended to use the day 5 embryonic culture medium (8).
Given the huge difference in the dimensions of these 2 chromosomes, it is quite clear that the extraction of the Y chromosome and the study of its genes is more difficult than the extraction and study of the genes of the X chromosome, particularly in the extraction of embryo's genetic material method from the culture medium where the amount of DNA obtained for examination is also little.
Our finding is consistent with the study that showed that the amount of DNA obtained from culture media was not sufficient for detecting accurately the testis-specific protein Y-Linked 1 gene, which is located on the Y chromosome but was enough for detecting the TBC1 domain family member 3 gene, which is located on chromosome 17 (12).
- Conclusion
Acknowledgements
This work was supported by grants from the Infertility Center, Imam Khomeini hospital, Ahvaz Jundishapur University of Medical Sciences, Khuzestan, Iran.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Type of Study: Original Article |
Subject:
Perinatology
References
1. Basille C, Frydman R, El Aly A, Hesters L, Fanchin R, Tachdjian G, et al. Preimplantation genetic diagnosis: State of the art. Eur J Obstet Gynecol Reprod Biol 2009; 145: 9-13. [DOI:10.1016/j.ejogrb.2009.04.004] [PMID]
2. Tasca RJ, McClure ME. The emerging technology and application of preimplantation genetic diagnosis. J Law Med Ethics 1998; 26: 7-16. [DOI:10.1111/j.1748-720X.1998.tb01901.x] [PMID]
3. Baruch S, Javitt G, Scott J, Hudson K. Preimplantation genetic diagnosis: A discussion of challenges, concerns, and preliminary policy options related to the genetic testing of human embryos. USA: Genetics and Public Policy Center; 2004.
4. Harper JC, SenGupta SB. Preimplantation genetic diagnosis: State of the art. Hum Genet 2012; 131: 175-186. [DOI:10.1007/s00439-011-1056-z] [PMID]
5. Handyside AH, Kontogianni EH, Hardy K, Winston RM. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 1990; 344: 768-770. [DOI:10.1038/344768a0] [PMID]
6. Handyside AH, Penketh R, Winston R, Pattinson J, Delhanty J, Tuddenham E. Biopsy of human preimplantation embryos and sexing by DNA amplification. Lancet 1989; 333: 347-349. [DOI:10.1016/S0140-6736(89)91723-6]
7. Li P, Song Z, Yao Y, Huang T, Mao R, Huang J, et al. Preimplantation genetic screening with spent culture medium/blastocoel fluid for in vitro fertilization. Sci Rep 2018; 8: 1-10. [DOI:10.1038/s41598-018-27367-4] [PMID] [PMCID]
8. Yang L, Lv Q, Chen W, Sun J, Wu Y, Wang Y, et al. Presence of embryonic DNA in culture medium. Oncotarget 2017; 8: 67805-67809. [DOI:10.18632/oncotarget.18852] [PMID] [PMCID]
9. Kuznyetsov V, Madjunkova S, Antes R, Abramov R, Motamedi G, Ibarrientos Z, et al. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS One 2018; 13: e0197262. [DOI:10.1371/journal.pone.0197262] [PMID] [PMCID]
10. Petersen CG, Mauri AL, Vagnini LD, Renzi A, Petersen B, Matilla MC, et al. Randomized comparison of two commercial culture media (Cook and Vitrolife) for embryo culture after IMSI. JBRA Assist Reprod 2019; 23: 33-36. [DOI:10.5935/1518-0557.20180058] [PMID] [PMCID]
11. Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, Peinado V, et al. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum Reprod 2018; 33: 745-756. [DOI:10.1093/humrep/dey028] [PMID]
12. Galluzzi L, Palini S, De Stefani S, Andreoni F, Primiterra M, Diotallevi A, et al. Extracellular embryo genomic DNA and its potential for genotyping applications. Future Sci OA 2015; 1: FSO62. [DOI:10.4155/fso.15.62] [PMID] [PMCID]
13. Ethics Committee of the American Society of Reproductive Medicine. Sex selection and preimplantation genetic diagnosis. Fertil Steril 2004; 82 (Suppl.): S245-S248.
14. Rajan-Babu I-Sh, Lian M, Cheah FSH, Chen M, Tan ASC, Prasath EB, et al. FMR1 CGG repeat expansion mutation detection and linked haplotype analysis for reliable and accurate preimplantation genetic diagnosis of fragile X syndrome. Expert Rev Mol Med 2017; 19: e10. [DOI:10.1017/erm.2017.10] [PMID] [PMCID]
15. Handyside AH, Lesko JG, Tarín JJ, Winston RM, Hughes MR. Birth of a normal girl after in vitro fertilization and preimplantation diagnostic testing for cystic fibrosis. N Engl J Med 1992; 327: 905-909. [DOI:10.1056/NEJM199209243271301] [PMID]
16. Meseguer M, Garrido N, Remohí J, Simón C, Pellicer A. Gender selection: Ethical, scientific, legal, and practical issues. J Assist Reprod Genet 2002; 19: 443-446. [DOI:10.1023/A:1016876026359] [PMID] [PMCID]
17. Joshi BD, De R, Goyal SP. Utility and applicability of a universal set of primers in identifying the sex of South and Southeast Asian mammals. Zool Stud 2019; 58: e19.
18. Osman J, Murad NAA, Chin SF, Jamal R. Highly sensitive and reliable human sex determination using multiplex PCR. Asia-Pacific J Mol Med 2016; 4: 1.
19. Settin A, Elsobky E, Hammad A, Al-Erany A. Rapid sex determination using PCR technique compared to classic cytogenetics. Int J Health Sci 2008; 2: 49-52.
20. Wu H, Ding C, Shen X, Wang J, Li R, Cai B, et al. Medium-based noninvasive preimplantation genetic diagnosis for human α-thalassemias-SEA. Medicine 2015; 94: e669. [DOI:10.1097/MD.0000000000000669] [PMID] [PMCID]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








