Wed, Jul 16, 2025
[Archive]
Volume 22, Issue 2 (Febraory 2024)
IJRM 2024, 22(2): 127-136 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Farajnezhad N, Ghandil P, Tahmasebi-Birgani M, Mohammadi-Asl J. Folate gene polymorphisms CBS 844ins68 and RFC1 A80G and risk of Down syndrome offspring in young Iranian women: A cross-sectional study. IJRM 2024; 22 (2) :127-136
URL: http://ijrm.ir/article-1-3082-en.html
URL: http://ijrm.ir/article-1-3082-en.html
1- Department of Medical Genetics, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
2- Department of Medical Genetics, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Cellular and Molecular Research Center, Medical Basic Sciences Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. ,tahmasebi.birgani62@gmail.com
3- Noorgene Genetics Laboratory, Ahvaz, Iran.
2- Department of Medical Genetics, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Cellular and Molecular Research Center, Medical Basic Sciences Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. ,
3- Noorgene Genetics Laboratory, Ahvaz, Iran.
Full-Text [PDF 1180 kb]
(347 Downloads)
| Abstract (HTML) (629 Views)
Full-Text: (131 Views)
1. Introduction
One of the most prevalent chromosomal causes of intellectual disability has been connected to Down syndrome (DS) (OMIMI # 190685). The syndrome is characterized by full or partial trisomy of chromosome 21. The incidence is approximately 1 in 1000 newborns and varies among the different populations (1). Full trisomy 21 is mainly caused by chromosomal nondisjunction during maternal meiosis-I. Free trisomy 21 is a sporadic event- if recurrence occurs, the gonadal mosaicism and Robertsonian translocation should be checked. Advanced maternal age (≥ 35 yr) is a well-approved risk factor for DS. However, several women < 35 yr at conception are predisposed to chromosomal nondisjunction and DS babies (2).
Folate is a natural form of vitamin B9, playing a vital role in DNA synthesis and methylation. In 1999, irregular folate metabolism was suggested to increase the probability of chromosome 21 nondisjunction through impairing the methylation levels in the pericentromeric regions of chromosome 21 and triggering chromosomal nondisjunction and genomic instability, (3, 4). Various studies considered the association between maternal folate gene polymorphisms and increased risk of DS; however, the results are controversial and remains questionable whether these polymorphisms in the mother are causal in determining DS susceptibility in the offspring (4, 5).
Here, 2 of the well-known folate genes are considered. The folate transporter reduced folate carrier protein (RFC1) on chromosome 21, mediates folate uptake into the cells. The polymorphism A80G (rs1051266) is the most prevalent variant in the RFC1 gene replacing the amino acid histidine with arginine in the exon-2 and decreasing the plasma levels of folate and homocysteine. A positive association between A80G and DS was documented in different populations (6). Cystathionine beta-synthase (CBS) is another vital enzyme in folate metabolism and catalyzes the conversion of homocysteine and serine to cystathionine. Polymorphism c.844ins68 is a frequent polymorphism of the CBS gene that results in an insertion of a 68-bp within exon 8 and duplicates the intron 7 acceptor splice site, which may lead to 2 CBS mRNA variants through alternative splicing. Previously published studies have documented that the CBS 844ins68 polymorphism may be related to DS (7).
The goal of this study was to see if polymorphisms CBS 844ins68 and RFC1 A80G may be linked to an increased risk of having DS among young Iranian mothers and used as maternal risk factor for DS. Although several researches on the function of Hcy/folate metabolism in the etiology of DS has been published in different geographical regions and populations, bringing both supportive data and innovative understandings, no study has existed on these polymorphisms in the Iranian population. The genetic counseling of DS would benefit significantly if the RFC1 A80G and CBS 844in68 polymorphisms could be associated to an increased risk of developing DS in females. Recent studies have suggested that DS children are more likely to be born from young mothers (8), we selected mothers whose age was under 30 yr at the time of pregnancy with DS babies. Moreover, we tried to reduce the effect of maternal age on the incidence of DS.
Therefore, in mothers of DS children, the syndrome may have arisen from something other than maternal age. To our knowledge, the present study is a basic study whose data can be useful in revealing the molecular mechanisms of the process of chromosome nondisjunction, and its goal is not to reach a diagnostic method and an alternative to conventional DS screening systems.
2. Materials and Methods
This cross-sectional study was conducted on 200 participants from June 2019 to April 2021 in Iran. Genomic DNA was extracted from the peripheral blood of 100 DS mothers (DSM) and 100 control mothers (CM). CBS 844ins68 was genotyped using polymerase chain reaction (PCR)-electrophoresis, while RFC1 A80G was analyzed through restriction fragment length polymorphism (RFLP).
2.1. Study population
To decrease the effect of maternal age, mothers older than 35 were excluded (n = 35) from this study. Then, 100 mothers, aged below 35 (n = 100), who had given birth to at least 1 child with DS, confirmed to have full trisomy 21 by karyotyping, were included in this study as DSM. Both mothers and fathers of children with DS having normal karyotype and carriers of translocation or iso chromosomes were excluded from this study. They were collected from June 2019 to April 2021 through medical genetics laboratories. 100 age- and ethnic-matched mothers whose children were not affected by DS were included in this project as CM. They had at least 2 healthy children and no history of miscarriage or abnormal pregnancies. All mothers belonged to the Khuzestan Province. The serum folate level of both DSM and CM were within the normal range. A questionnaire was used to collect data from mothers and all enrolled participants signed the informed consent.
2.2. Genomic DNA extraction
Genomic DNA was extracted from 5 mL of peripheral blood lymphocytes using a DNA extraction kit (Kiagene, Iran) according to its user protocol. Extracted DNAs were stored at -20°C until use.
2.3. Detection of polymorphisms
2.3.1. CBS 844ins68 genotyping
Since 844ins68 polymorphism is a 68-bp insertion in the CBS gene, genotyping was done using PCR and agarose gel electrophoresis. Specific forward and reverse primers were 5′-CGCCCTCTGCAGATCATTGG-3′ and 5′-CCTTCCACCTCGTAGGTTGTC-3′, respectively, to amplify a 98 bp amplicon of the CBS gene in wild-type. The PCR reaction was prepared in the total volume of 25 μl containing 1× reaction buffer, 0.25 mM of each dNTP, 2 pmol/l of each primer, 0.4 μg genomic DNA templates, and 1.5 U of Taq DNA polymerase. The PCR was set up in thermocycler as 5 min denaturation at 95°C for initial denaturation followed by 33 cycles of denaturation; 95°C (1 min), annealing; 62°C (45 sec), extension; 72°C (45 sec), final extension; 72°C (7 min). A negative control reaction was prepared in all PCR reactions. The 2% agarose gel electrophoresis was used to discriminate all possible genotypes. The expected electrophoretic pattern was a 98 bp fragment for the wild-type homozygous genotype (Ins-/-), 2 fragments of 98 bp and 166 bp for the heterozygous genotype (Ins+/-), and a fragment of 166 bp for CBS 844ins68 homozygous genotype (Ins+/+).
2.3.2. RFC1 A80G genotyping
Polymorphism RFC1 A80G genotype analyses were done using PCR-RFLP. Specific forward and reverse primers were 5′-AGTGTCACCTTCGTCCC-3′ and 5′-GTTCTTGAAGTGCGCCCT-3′, respectively. The PCR was set up in a thermocycler as 5 min denaturation at 95°C for initial denaturation followed by 33 cycles of denaturation; 95°C (1 min), annealing; 53°C (45 sec), extension; 72°C (45 sec), final extension; 72°C (7 min). The PCR products were then subjected to digestion with CfoI (Sigma) endonuclease for 3 hr. The expected RFLP pattern were 3 fragments of 125, 68, and 37 bp in the presence of the 80G allele, while the 80A allele produced 2 pieces of 162 and 68 bp.
2.4. Sanger DNA sequencing
The amplicon sequence was then read using Big Dye Terminators (Applied Bio systems™, Genetic Analyzer; Applied Biosystems, Foster City, CA, USA) to confirm the specificity of used primers and the accuracy of genotyping.
2.5. Study of gene-gene interactions and risk of DS
All the possible combinations between RFC1 A80G and CBS 844ins68 genotypes were calculated by a 2-by-4 table to measure the excess risk due to the presence of 2 A80G and 844ins68 polymorphisms genotypes in the same woman.
2.6. Ethical considerations
This study was ethically approved by Ahvaz Jundishapur University of Medical Sciences Research Affairs, Ahvaz, Iran (Ethical Code: IR.AJUMS.REC.1397.688) and was experimentally performed in department of Medical Genetics and Cellular and Molecular Research Center. Before starting the project, written informed consent was obtained from all individuals who participated in this project.
2.7. Statistical analysis
Statistical Package for the Social Sciences, version 22.0, SPSS Inc., Chicago, Illinois, USA (SPSS) and Chi-square test were used for data analyses. The Hardy-Weinberg equilibrium (HWE) was tested using allele and genotype frequencies to identify genotyping errors. The odds ratio (OR) and 95% confidence intervals (CIs) in DSM/CM were calculated among 4 frequent genetic inheritance models, including the Allelic model, dominant model, recessive model, and over-dominant model. The genotype can be a major allele homozygote (MM), a heterozygote (Mm) or a minor allele homozygote (mm). The dominant model compares heterozygote (Mm)+ minor allele homozygote (mm) vs. major allele homozygote (MM), recessive model compares the minor allele homozygote (mm) vs. heterozygote (Mm)+ major allele homozygote (MM), over-dominant Model compares heterozygote (Mm) vs. major allele homozygote (mm)+ minor allele homozygote (mm). Probabilistic Model selection was performed using the Akaike information criterion (AIC) value. The best-fit inheritance model with the smallest AIC was considered the inheritance Model. The p-value < 0.05 was considered significant.
3. Results
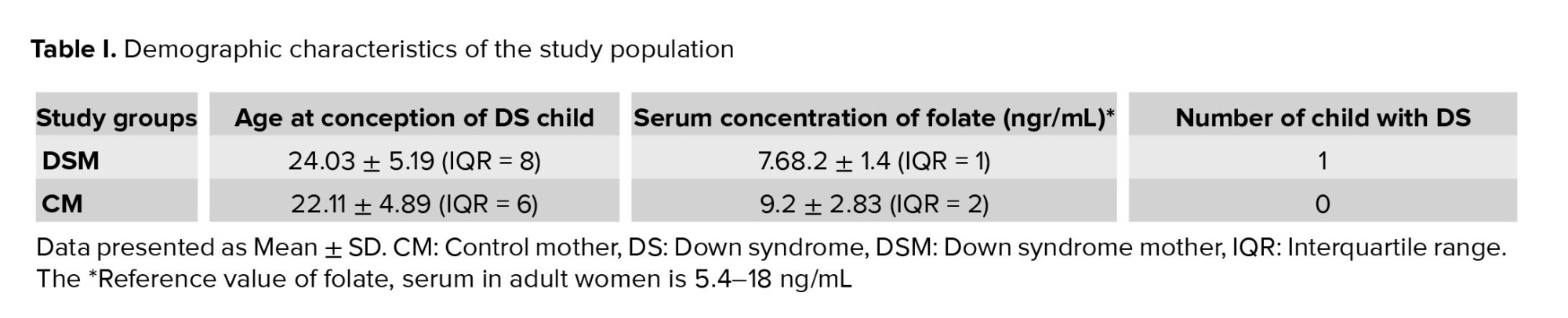
Table I shows the demographic characterization of the study population. In the group DSM, the mean age during the pregnancy of DS was 24.03 ± 5.19 yr, while the CM had a mean age of 22.11 ± 4.89. Serum folate level of both DSM and CM were within the normal range (6-10 ng/mL) and the difference was not significance (p > 0.05). The mean serum concentration of folate in DSM and CM were 7.68.2 ± 1.4 and 9.2 ± 2.83 ngr/mL respectively. In DSM group, each family had only 1 DS child who was not necessarily their first child. Parents of children with DS having normal karyotype and carriers of translocation or iso-chromosomes were excluded from this study.
3.1. Polymorphism RFC1 A80G was highly detected in DSM
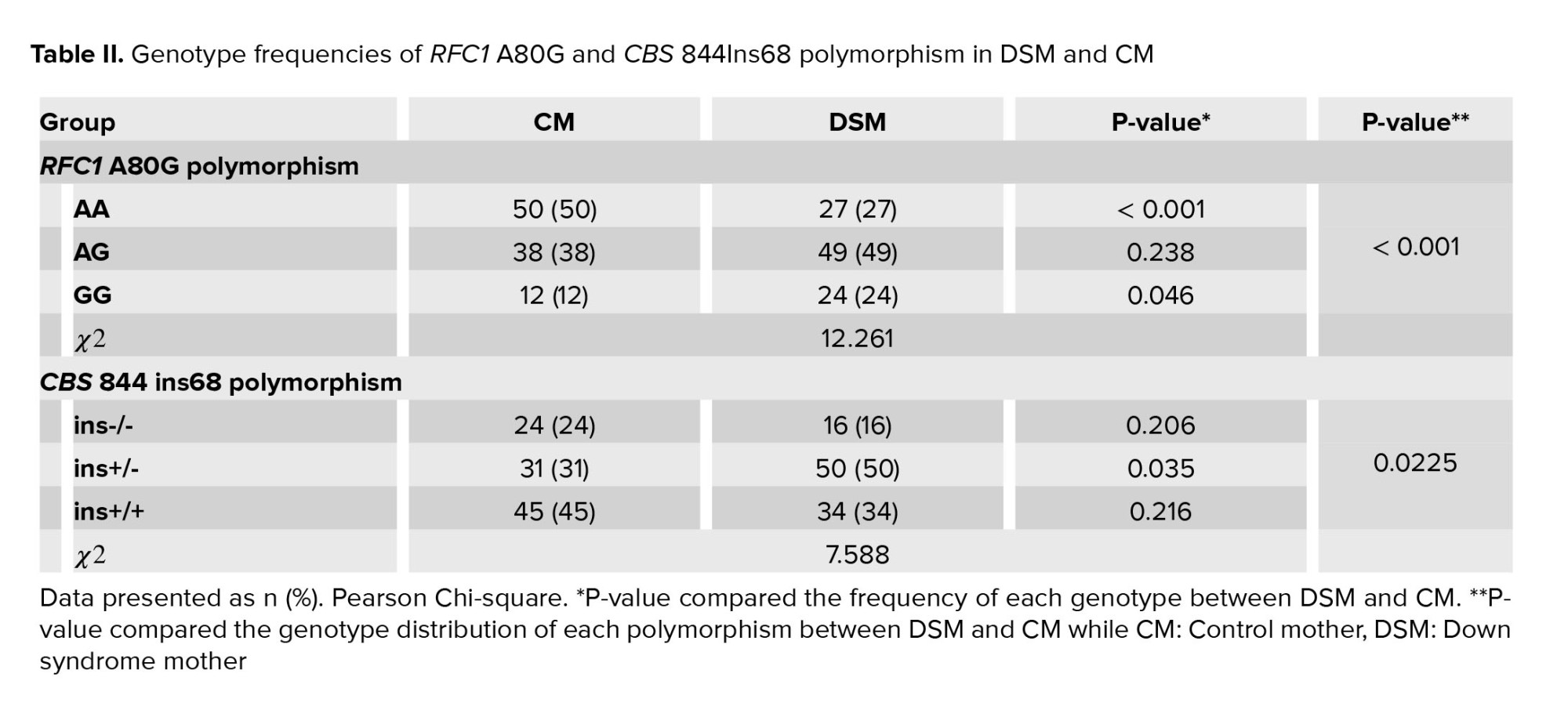
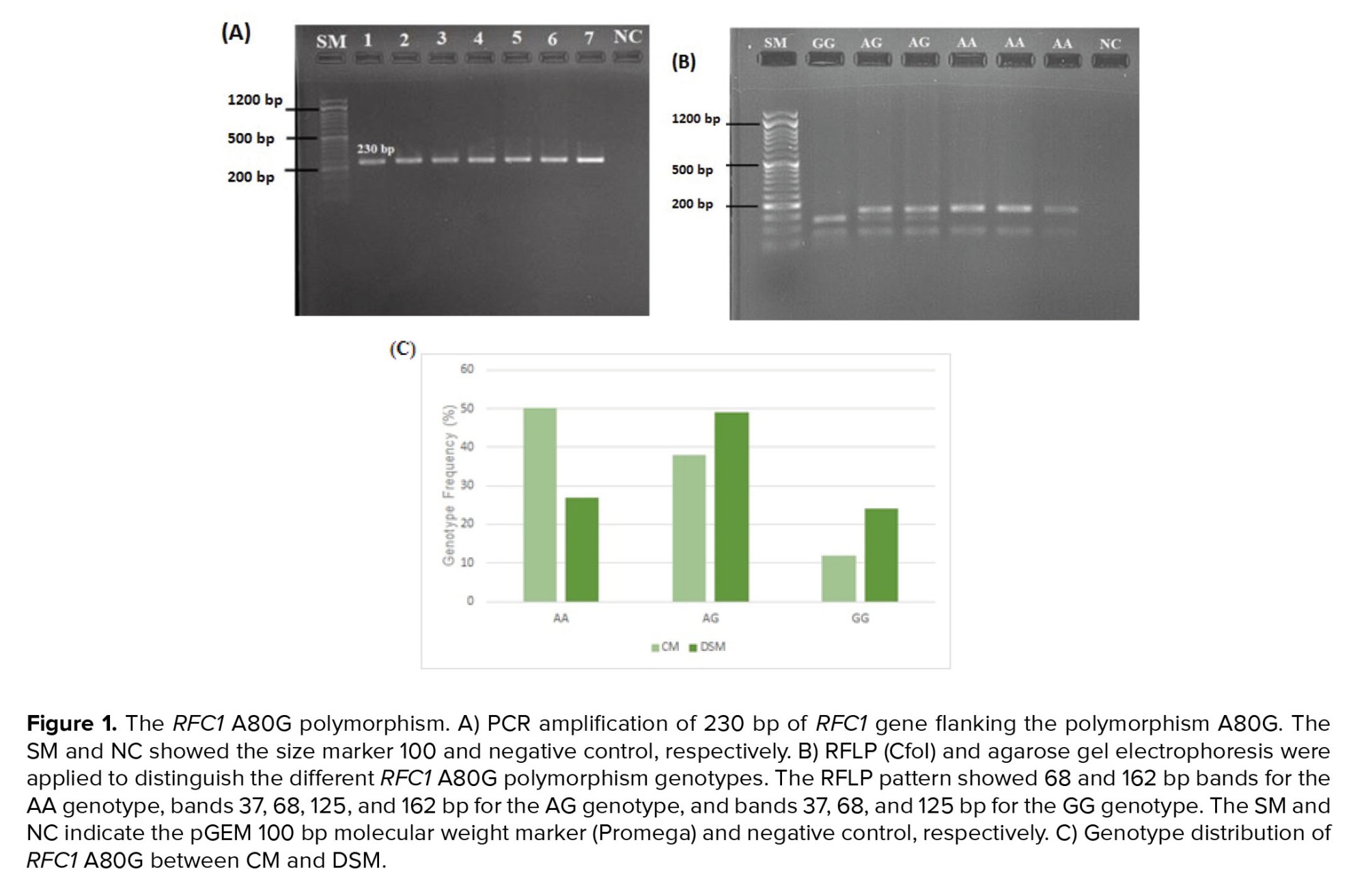
The genotype of RCF1 A80G polymorphism was evaluated through an RFLP assay. A 230 bp fragment flanking the A80G polymorphism was amplified during PCR (Figure 1A). Figure 1B shows the RFLP pattern of some participants, illustrating 68 and 162 bp bands for AA genotype, bands 37, 68, 125, and 162 bp for AG genotype, and bands 37, 68, and 125 bp for GG genotype. The genotype frequency of A80G was compared between DSM and CM (Figure 1C). As indicated, a significant difference was observed in genotype frequency of A80G between DSM and CM (χ2 = 12.261, p = 0.002) (Table II).
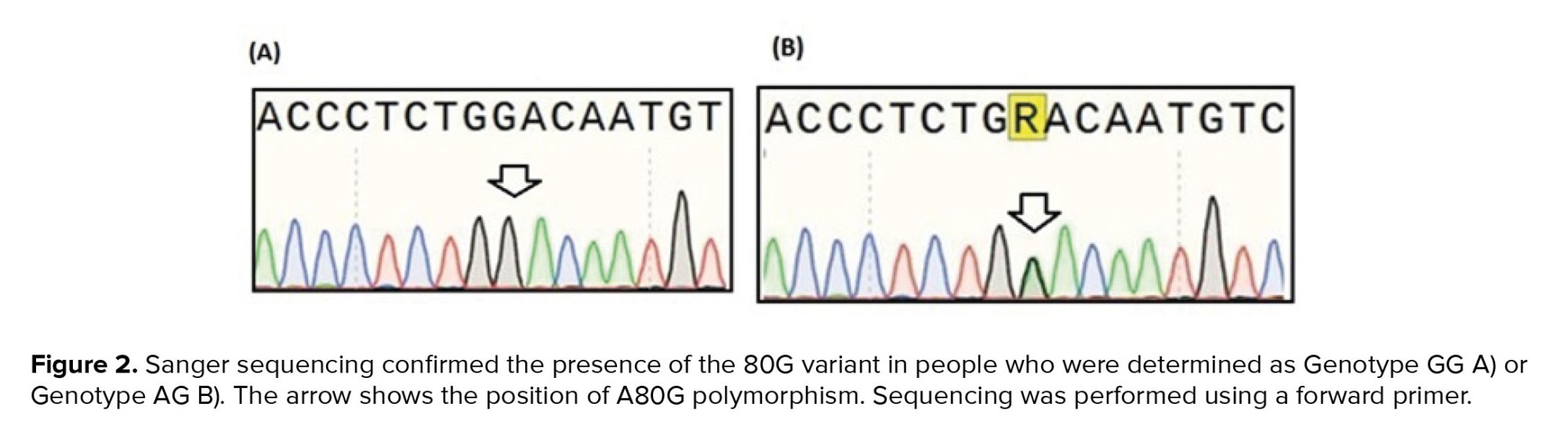
Figure 2 shows the results of Sanger sequencing of 2 heterozygous and homozygous participants for A80G. The distribution of all RFC1 A80G genotypes in our study population was consistent with the Hardy-Weinberg equilibrium (χ2 = 0.036, p = 0.82).
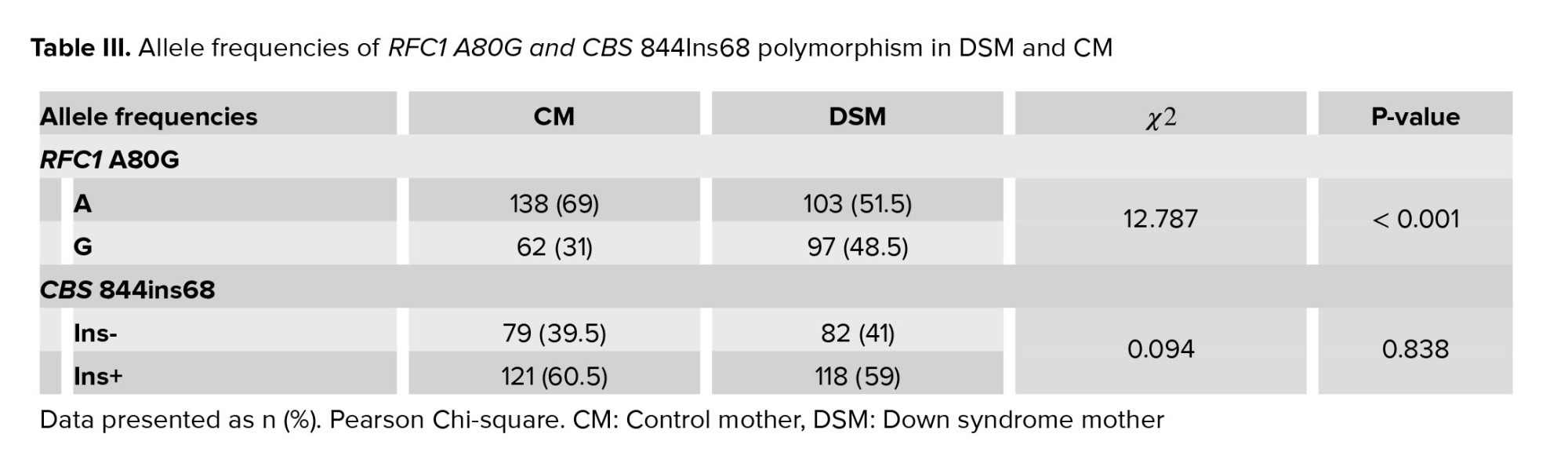
Table III compared the frequency of the “A” allele, which was found to be 69% in CM and 51.5% in DSM, While the “G” allele frequency was 31% and 48.5% among CM and DSM, respectively. A significant difference in RFC1 A80G allelic frequencies was found between DSM and CM (χ2 = 12.261, p < 0.001), and the frequency of the G allele was significantly higher in DSM than the CM (Table III). Table IV indicates the summary estimates for the OR of RFC1 A80G for the association between A80G and the risk of DS in allele, dominant, recessive, and over-dominant models. RFC1 A80G polymorphism was significantly correlated with having DS risk in both dominant and recessive inheritance comparison models (OR = 2.316, p = 0.03 for the recessive model, and OR = 2.704, p = 0.001 in dominant model). The dominant model is the best inheritance model as it yields the strongest association with the smallest AIC (-8.8). The minor G allele was associated with an increased risk of DS (OR = 2.09, 95% CI = 1.39-3.15, p < 0.001) (Table IV).
3.2. Polymorphism CBS 844in68 and increased risk of having DS baby in mothers
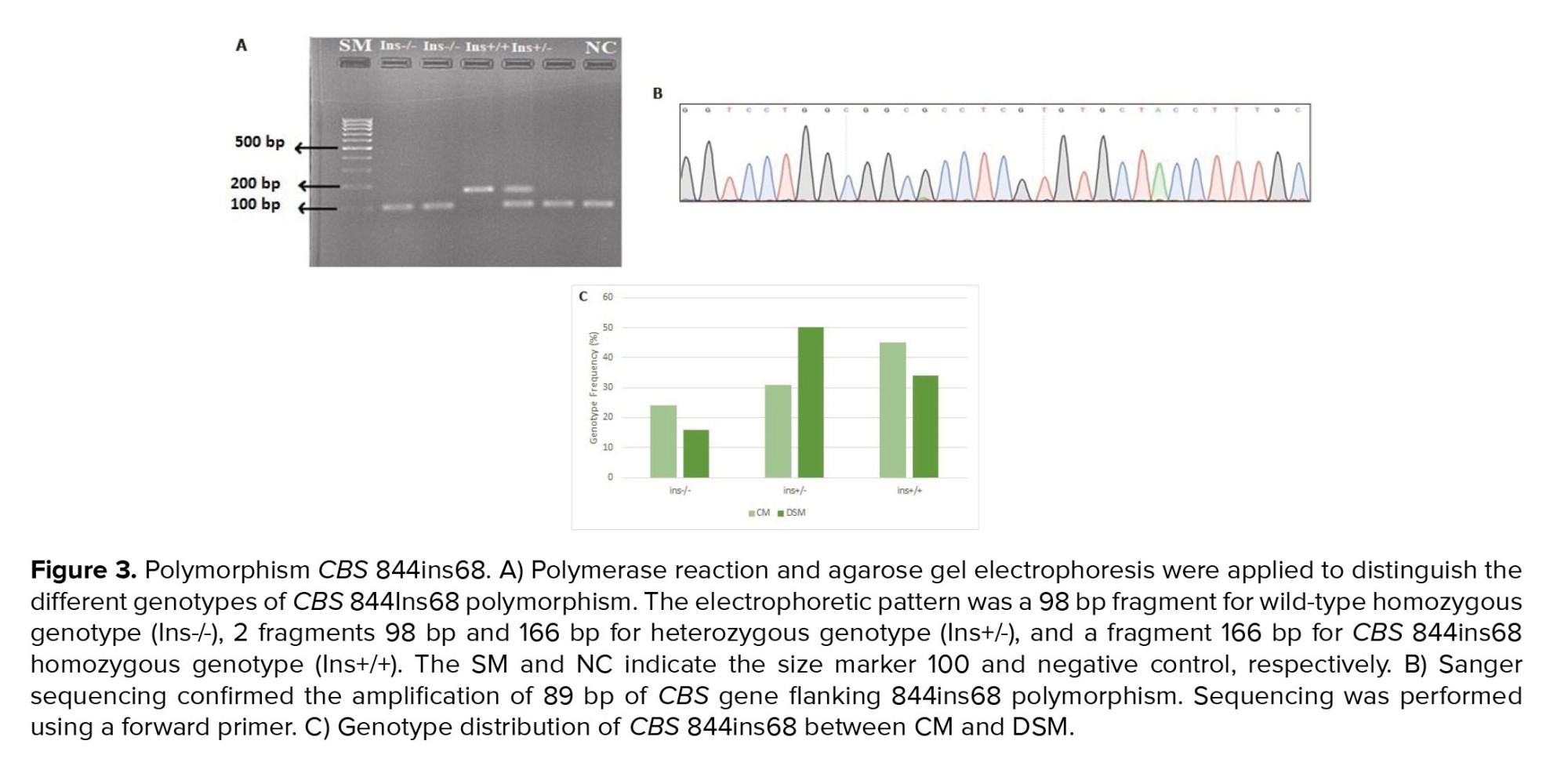
As indicated in the method section, polymorphism CBS 844ins68 was genotyped using PCR and electrophoresis. Figure 3A shows the result of agarose gel electrophoresis for some participants. As expected, a 98 bp fragment was observed for the wild-type homozygote genotype (Ins-/-), while 2 fragments of 98 bp and 166 bp were detected in the heterozygous genotype (Ins+/-). The presence of a 166 bp fragment confirmed the homozygous of (Ins+/Ins+).
The true amplification of the CBS 844ins68 flanking region was confirmed through the sanger sequencing (Figure 3B). The genotype frequency of 844ins68 was compared between DSM and CM (Figure 3C, Table II). As indicated, a significant difference was observed in genotype frequency of CBS 844ins68 between DSM and CM (χ2 = 7.588, p = 0.0225). The observed genotype frequency for the Ins -/- genotype in DSM was 16% compared to 24% in CM (p = 0.206); Ins+/- genotype frequency was 50% in DSM, and 31% in CM which was statistically significant (p = 0.035), and the Ins+/+ genotype was 34% in DSM compared to 45% in CM (p = 0.216). The genotype distribution of CBS 844Ins68 polymorphism significantly differed between CM and DSM (χ2 = 0.112, p < 0.001). The distribution of all CBS genotypes was inconsistent with HWE (χ2 = 12.261, p = 0.737) (Table II, Figure 3C). The allele frequency of 844ins68 is shown in table III. As indicated, the “Ins +” allele frequency was found to be 0.605 in CM and 0.59 in DSM (p = 0.838). The “ins-” allele frequency was 0.395 and 0.41 among CM and DSM, respectively. No significant difference was observed in SNP allelic frequencies of DSM and CM (p = 0.838) (Table III).
3.3. Gene-gene interaction between CBS 844ins68 and RFC1 A80G polymorphisms
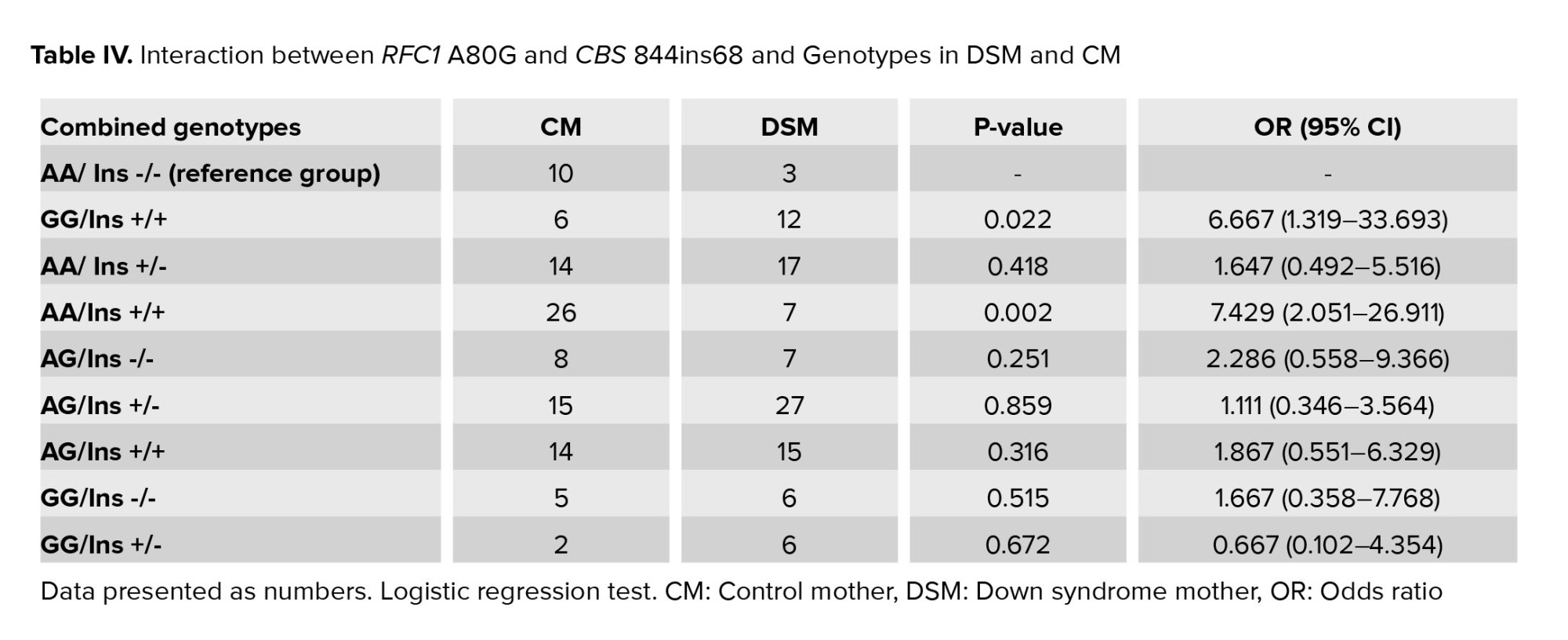
The interaction of CBS 844ins68 and RFC1 A80G in all 200 genotyped DSM and CM was summarized in table IV. As illustrated, a combination of both polymorphisms RFC1 A80G and CBS 844ins68 in a homozygous state (GG/Ins+/+) can significantly increase the risk of DS (OR = 6.667 and p = 0.022).
4. Discussion
This is the first study showing the positive association of CBS 844ins68 and RFC1 A80G polymorphism as a risk factor for DS in Iranian young DS mothers who were residents of southwest Iran.
We found that only the heterozygous state of CBS 844ins68 polymorphism was more frequent among DSM than the CMs and half of the DSM were positive for the 844ins variant. The calculated OR was 2.419, which means heterozygous mothers of the 844ins68 variant have around a 2.4-fold increase in the risk of DS than the mothers with other genotypes of 844ins68. The frequency of the 80G allele in DSM (48.5%) is significantly higher than the observed frequency in CM (31%). The OR was calculated as 2.09. We observed that 24% of DSM were homozygous for the 80G variant (GG genotype), and 49% were heterozygous for 80G (AG genotype). In CM, the frequency of GG and AG genotypes were 12% and 38%, respectively.
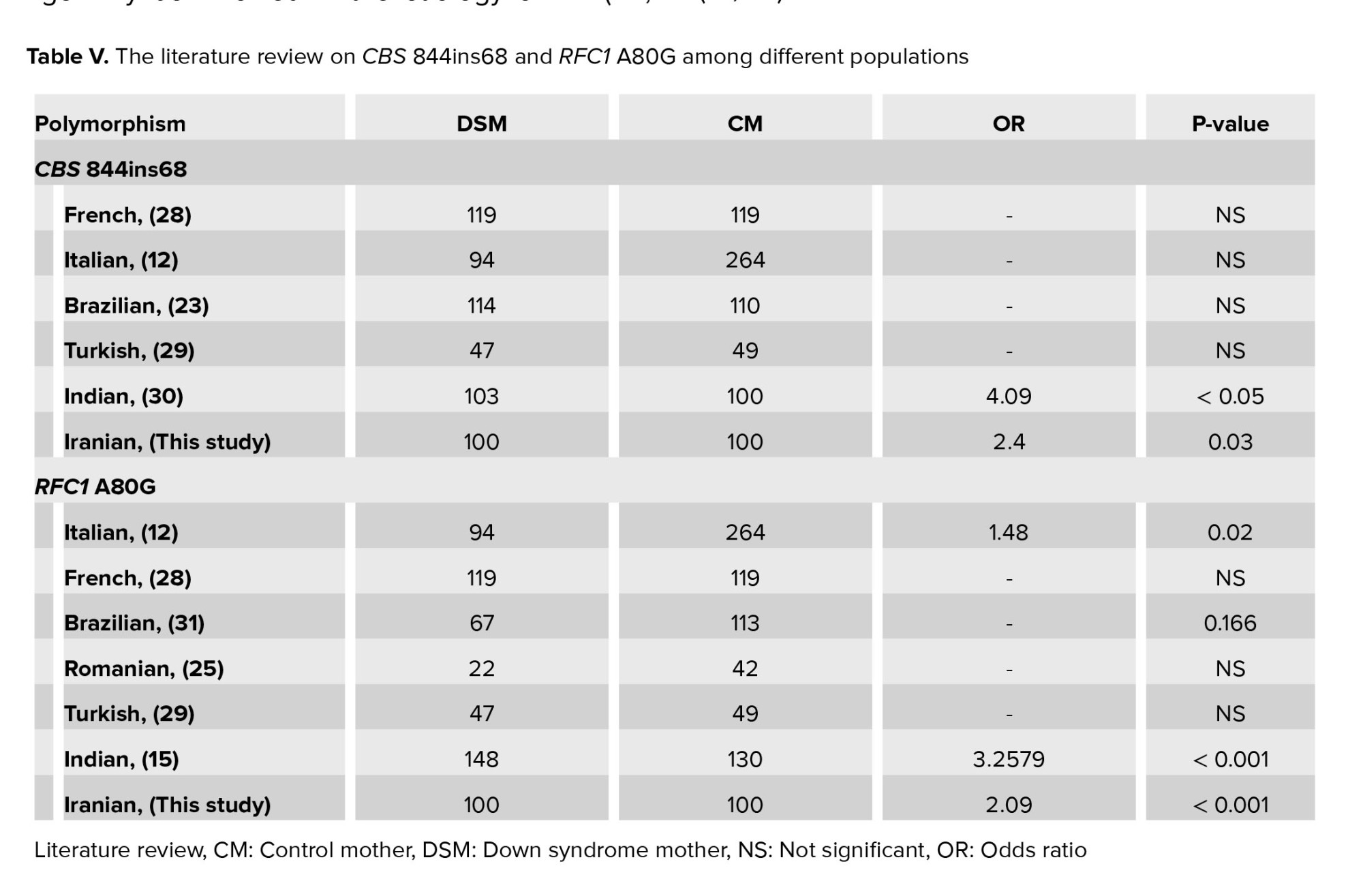
The gene RFC1 encodes a transporter of folate into and out of cells. Mutation in the RFC1 gene can be associated with multiple defects in mice models of birth defect, and supplementation with folic acid can increase the survival of embryos (9). Through HPA RNA-seq normal tissues, a high expression level of RFC1 was found in the liver, placenta, heart, and whole blood (https://www.ncbi.nlm.nih.gov/gene/6573). RFC1 has not been directly connected to the increased level of total homocysteine, but it may decrease the availability of folic acid by the developing fetus, resulting in growth problems. Different variants of RFC1 can influence the concentration of folate metabolism (10). In the latest release of dbSNP (https://www.ncbi.nlm.nih.gov/snp/), 3027 single nucleotide variants have been reported in the RFC1 gene. However, polymorphism A80G is the most frequent nucleotide change in the RFC1 gene. The A80G is a missense variant resulting in the substitution of histidine amino acid codon (CAG) to arginine (CGG) and altered concentrations of folate metabolic metabolism products (6). Chango et al. reported that the A80G variant of this gene could increase the total homocysteine level in the homozygous GG state (10). Yet, there appears to be little or no consensus about how to the allele inherited and further analysis is needed. However, it is well documented that the transporter protein derived from the RFC1 allele 80G is less functional than the protein encoded by the wild-type allele (80A). It has been documented that women who are homozygous of RFC1 80G have a 2-fold higher risk of having a baby with a birth defect than those with the 80A variant (11). Totally, 6 studies investigated the association of RFC1 A80G polymorphism as a maternal genetic risk factor for DS showing the positive association among Italian, French, Romanian, Turkish, Indian, and Iranian mothers as well (Table V).
Similar to Iranian mothers, the Italian and Indian populations showed a positive association between the presence of RFC1 A80G polymorphism and increased risk of DS. However, no association was observed in a population of Turkish, French, Brazilian, and Romanian. The frequency of the 80G variant in Italian mothers was 49.5%, close to our observed frequency of 48.5% among Iranian mothers (12). This finding was unsurprising as genetic similarity and close relationship between Iranians and Italians based on previously published studies. Iranian-related ancestry was evidenced in Italy's Sardinia and Sicily cities (13, 14). The frequency of the 80G allele was 37.16 among Indian mothers of DS (15). The same story was documented among the Iranian and Indian populations due to the migratory flux between these countries (16). In the case of a population with a lack of association, the unlike results may arise from the difference in their nutritional regime or distinct genetic context.
The CBS, a crucial enzyme in the folic acid cycle, is strongly expressed in normal brain tissues and the liver, pancreas, and kidney. The 844ins68 (Allele ID: 231132) is the most common polymorphism of the CBS gene, which involves a 68-bp insertion in the coding region of exon8 at position 844. It causes a frameshift, introducing 2 nonsense codons in the same exon. The polymorphism duplicates the intron-exon boundary of exon 8, producing an alternative splicing site (17). In 1995, the CBS 844ins68 was first reported in a homocystinuria individuals (18). The current study showed a significant difference in the genotype frequency of CBS 844ins68 between DSM and CM. The “Ins+” allele frequency was low (0.605 in CM and 0.59 in DSM. Extensive ethnic and geographic variability was connected with CBS 844ins68 polymorphism (19, 20) (Table V).
The frequency of the heterozygous genotype of the 844ins68 allele is around 11.7% in normal North American populations; the Netherlands, Spain, the United States, and Ethiopia have an average prevalence of 11-17%, and the population of China, Czechoslovakia, India, Chile, and Colombia have a low prevalence of 1-10% for heterozygous genotype. The 844ins68 allele has not been reported in Indonesia or Japan (21).
Similar to our result, in a study on the frequency of 844ins68 in Southern Iran, this population showed great similarity with other Caucasian populations, especially in South Italy and North America. In contrast, it varied from East Asian and African populations (22). Although the 844ins68 does not seem to disturb the activity of the CBS enzyme, the frequency is increased in people with premature coronary-artery disease (18). The association of CBS 844ins68 with DS has been studied in different populations (Table V), but the results were controversial. Fintelman-Rodrigues et al. in a study on the Brazilian people, showed that CBS 844Ins68 polymorphism did not correlate with DS pregnancies. While the combined genotype of this polymorphism and other polymorphisms of the folic acid cycle, such as RFC1 80G>A, MTRR 66A>G, MTR 2756A>G, MTHFR 1298A>C were associated with the increased risk of having DS babies (23). However, Scala et al. suggested that CBS 844Ins68 could not increase the risk of having a baby with DS in Italy (12). Such contradictory results may relate to various factors, including sample size or differences in geographical area and racial background, which should be considered in future studies. We also found that the combination of both RFC1 GG genotypes with CBS 844ins +/+ can increase the risk of having DS babies to 6.667.
One of the valuable points of this study was the evaluation of RFC1 A80G and CBS 844ins68 in 100 young mothers under 35. It has been shown that most DS children have been born to young women, triggering the idea that other risk factors than increased maternal age may be involved in the etiology of DS (24, 25). In 2009, Migliore and Migheli showed that chromosomal nondisjunction and missegregation are multifactorial traits presented by genetics and environment. They found that young mothers of DS children might have a genetic predisposition (like special polymorphisms) to abnormal chromosome segregation in somatic and germinal cells (26). In this study, we selected mothers with a mean maternal age of 24.03 ± 5.19 yr during the pregnancy of DS babies and excluded those over the age of 35 during the pregnancy. In fact, by considering such criteria, we reduced the effect of maternal age on the incidence of DS. Therefore, in mothers of DS children, the syndrome may have arisen from something other than maternal age. Of note, this idea has not been considered in most of the studies that estimated the association of maternal polymorphisms with having DS babies (15, 27).
5. Conclusion
In conclusion, this study showed that RFC1 A80G and CBS 844Ins68 are 2 polymorphisms of folate metabolism that can be involved in the etiology of DS among Iranian mothers of Khuzestan province.
Data availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author contributions
MTB and NF had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: MTB. Acquisition, analysis, or interpretation of data: MTB, NF, PGH and JMA. Drafting of the manuscript: NF, MTB and PGH. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: NF and MTB. Supervision: MTB and PGH.
Acknowledgments
The authors appreciate the efforts of all colleagues at Ahvaz Jundishapur University of Medical Sciences Research Affairs, Department of Medical Genetics and Cellular and Molecular Research Center for their generous assistance during this project (Grant No: CMRC-9717).
Conflict of Interest
The authors declare that there is no conflict of interest.
One of the most prevalent chromosomal causes of intellectual disability has been connected to Down syndrome (DS) (OMIMI # 190685). The syndrome is characterized by full or partial trisomy of chromosome 21. The incidence is approximately 1 in 1000 newborns and varies among the different populations (1). Full trisomy 21 is mainly caused by chromosomal nondisjunction during maternal meiosis-I. Free trisomy 21 is a sporadic event- if recurrence occurs, the gonadal mosaicism and Robertsonian translocation should be checked. Advanced maternal age (≥ 35 yr) is a well-approved risk factor for DS. However, several women < 35 yr at conception are predisposed to chromosomal nondisjunction and DS babies (2).
Folate is a natural form of vitamin B9, playing a vital role in DNA synthesis and methylation. In 1999, irregular folate metabolism was suggested to increase the probability of chromosome 21 nondisjunction through impairing the methylation levels in the pericentromeric regions of chromosome 21 and triggering chromosomal nondisjunction and genomic instability, (3, 4). Various studies considered the association between maternal folate gene polymorphisms and increased risk of DS; however, the results are controversial and remains questionable whether these polymorphisms in the mother are causal in determining DS susceptibility in the offspring (4, 5).
Here, 2 of the well-known folate genes are considered. The folate transporter reduced folate carrier protein (RFC1) on chromosome 21, mediates folate uptake into the cells. The polymorphism A80G (rs1051266) is the most prevalent variant in the RFC1 gene replacing the amino acid histidine with arginine in the exon-2 and decreasing the plasma levels of folate and homocysteine. A positive association between A80G and DS was documented in different populations (6). Cystathionine beta-synthase (CBS) is another vital enzyme in folate metabolism and catalyzes the conversion of homocysteine and serine to cystathionine. Polymorphism c.844ins68 is a frequent polymorphism of the CBS gene that results in an insertion of a 68-bp within exon 8 and duplicates the intron 7 acceptor splice site, which may lead to 2 CBS mRNA variants through alternative splicing. Previously published studies have documented that the CBS 844ins68 polymorphism may be related to DS (7).
The goal of this study was to see if polymorphisms CBS 844ins68 and RFC1 A80G may be linked to an increased risk of having DS among young Iranian mothers and used as maternal risk factor for DS. Although several researches on the function of Hcy/folate metabolism in the etiology of DS has been published in different geographical regions and populations, bringing both supportive data and innovative understandings, no study has existed on these polymorphisms in the Iranian population. The genetic counseling of DS would benefit significantly if the RFC1 A80G and CBS 844in68 polymorphisms could be associated to an increased risk of developing DS in females. Recent studies have suggested that DS children are more likely to be born from young mothers (8), we selected mothers whose age was under 30 yr at the time of pregnancy with DS babies. Moreover, we tried to reduce the effect of maternal age on the incidence of DS.
Therefore, in mothers of DS children, the syndrome may have arisen from something other than maternal age. To our knowledge, the present study is a basic study whose data can be useful in revealing the molecular mechanisms of the process of chromosome nondisjunction, and its goal is not to reach a diagnostic method and an alternative to conventional DS screening systems.
2. Materials and Methods
This cross-sectional study was conducted on 200 participants from June 2019 to April 2021 in Iran. Genomic DNA was extracted from the peripheral blood of 100 DS mothers (DSM) and 100 control mothers (CM). CBS 844ins68 was genotyped using polymerase chain reaction (PCR)-electrophoresis, while RFC1 A80G was analyzed through restriction fragment length polymorphism (RFLP).
2.1. Study population
To decrease the effect of maternal age, mothers older than 35 were excluded (n = 35) from this study. Then, 100 mothers, aged below 35 (n = 100), who had given birth to at least 1 child with DS, confirmed to have full trisomy 21 by karyotyping, were included in this study as DSM. Both mothers and fathers of children with DS having normal karyotype and carriers of translocation or iso chromosomes were excluded from this study. They were collected from June 2019 to April 2021 through medical genetics laboratories. 100 age- and ethnic-matched mothers whose children were not affected by DS were included in this project as CM. They had at least 2 healthy children and no history of miscarriage or abnormal pregnancies. All mothers belonged to the Khuzestan Province. The serum folate level of both DSM and CM were within the normal range. A questionnaire was used to collect data from mothers and all enrolled participants signed the informed consent.
2.2. Genomic DNA extraction
Genomic DNA was extracted from 5 mL of peripheral blood lymphocytes using a DNA extraction kit (Kiagene, Iran) according to its user protocol. Extracted DNAs were stored at -20°C until use.
2.3. Detection of polymorphisms
2.3.1. CBS 844ins68 genotyping
Since 844ins68 polymorphism is a 68-bp insertion in the CBS gene, genotyping was done using PCR and agarose gel electrophoresis. Specific forward and reverse primers were 5′-CGCCCTCTGCAGATCATTGG-3′ and 5′-CCTTCCACCTCGTAGGTTGTC-3′, respectively, to amplify a 98 bp amplicon of the CBS gene in wild-type. The PCR reaction was prepared in the total volume of 25 μl containing 1× reaction buffer, 0.25 mM of each dNTP, 2 pmol/l of each primer, 0.4 μg genomic DNA templates, and 1.5 U of Taq DNA polymerase. The PCR was set up in thermocycler as 5 min denaturation at 95°C for initial denaturation followed by 33 cycles of denaturation; 95°C (1 min), annealing; 62°C (45 sec), extension; 72°C (45 sec), final extension; 72°C (7 min). A negative control reaction was prepared in all PCR reactions. The 2% agarose gel electrophoresis was used to discriminate all possible genotypes. The expected electrophoretic pattern was a 98 bp fragment for the wild-type homozygous genotype (Ins-/-), 2 fragments of 98 bp and 166 bp for the heterozygous genotype (Ins+/-), and a fragment of 166 bp for CBS 844ins68 homozygous genotype (Ins+/+).
2.3.2. RFC1 A80G genotyping
Polymorphism RFC1 A80G genotype analyses were done using PCR-RFLP. Specific forward and reverse primers were 5′-AGTGTCACCTTCGTCCC-3′ and 5′-GTTCTTGAAGTGCGCCCT-3′, respectively. The PCR was set up in a thermocycler as 5 min denaturation at 95°C for initial denaturation followed by 33 cycles of denaturation; 95°C (1 min), annealing; 53°C (45 sec), extension; 72°C (45 sec), final extension; 72°C (7 min). The PCR products were then subjected to digestion with CfoI (Sigma) endonuclease for 3 hr. The expected RFLP pattern were 3 fragments of 125, 68, and 37 bp in the presence of the 80G allele, while the 80A allele produced 2 pieces of 162 and 68 bp.
2.4. Sanger DNA sequencing
The amplicon sequence was then read using Big Dye Terminators (Applied Bio systems™, Genetic Analyzer; Applied Biosystems, Foster City, CA, USA) to confirm the specificity of used primers and the accuracy of genotyping.
2.5. Study of gene-gene interactions and risk of DS
All the possible combinations between RFC1 A80G and CBS 844ins68 genotypes were calculated by a 2-by-4 table to measure the excess risk due to the presence of 2 A80G and 844ins68 polymorphisms genotypes in the same woman.
2.6. Ethical considerations
This study was ethically approved by Ahvaz Jundishapur University of Medical Sciences Research Affairs, Ahvaz, Iran (Ethical Code: IR.AJUMS.REC.1397.688) and was experimentally performed in department of Medical Genetics and Cellular and Molecular Research Center. Before starting the project, written informed consent was obtained from all individuals who participated in this project.
2.7. Statistical analysis
Statistical Package for the Social Sciences, version 22.0, SPSS Inc., Chicago, Illinois, USA (SPSS) and Chi-square test were used for data analyses. The Hardy-Weinberg equilibrium (HWE) was tested using allele and genotype frequencies to identify genotyping errors. The odds ratio (OR) and 95% confidence intervals (CIs) in DSM/CM were calculated among 4 frequent genetic inheritance models, including the Allelic model, dominant model, recessive model, and over-dominant model. The genotype can be a major allele homozygote (MM), a heterozygote (Mm) or a minor allele homozygote (mm). The dominant model compares heterozygote (Mm)+ minor allele homozygote (mm) vs. major allele homozygote (MM), recessive model compares the minor allele homozygote (mm) vs. heterozygote (Mm)+ major allele homozygote (MM), over-dominant Model compares heterozygote (Mm) vs. major allele homozygote (mm)+ minor allele homozygote (mm). Probabilistic Model selection was performed using the Akaike information criterion (AIC) value. The best-fit inheritance model with the smallest AIC was considered the inheritance Model. The p-value < 0.05 was considered significant.
3. Results
Table I shows the demographic characterization of the study population. In the group DSM, the mean age during the pregnancy of DS was 24.03 ± 5.19 yr, while the CM had a mean age of 22.11 ± 4.89. Serum folate level of both DSM and CM were within the normal range (6-10 ng/mL) and the difference was not significance (p > 0.05). The mean serum concentration of folate in DSM and CM were 7.68.2 ± 1.4 and 9.2 ± 2.83 ngr/mL respectively. In DSM group, each family had only 1 DS child who was not necessarily their first child. Parents of children with DS having normal karyotype and carriers of translocation or iso-chromosomes were excluded from this study.
3.1. Polymorphism RFC1 A80G was highly detected in DSM
The genotype of RCF1 A80G polymorphism was evaluated through an RFLP assay. A 230 bp fragment flanking the A80G polymorphism was amplified during PCR (Figure 1A). Figure 1B shows the RFLP pattern of some participants, illustrating 68 and 162 bp bands for AA genotype, bands 37, 68, 125, and 162 bp for AG genotype, and bands 37, 68, and 125 bp for GG genotype. The genotype frequency of A80G was compared between DSM and CM (Figure 1C). As indicated, a significant difference was observed in genotype frequency of A80G between DSM and CM (χ2 = 12.261, p = 0.002) (Table II).
Figure 2 shows the results of Sanger sequencing of 2 heterozygous and homozygous participants for A80G. The distribution of all RFC1 A80G genotypes in our study population was consistent with the Hardy-Weinberg equilibrium (χ2 = 0.036, p = 0.82).
Table III compared the frequency of the “A” allele, which was found to be 69% in CM and 51.5% in DSM, While the “G” allele frequency was 31% and 48.5% among CM and DSM, respectively. A significant difference in RFC1 A80G allelic frequencies was found between DSM and CM (χ2 = 12.261, p < 0.001), and the frequency of the G allele was significantly higher in DSM than the CM (Table III). Table IV indicates the summary estimates for the OR of RFC1 A80G for the association between A80G and the risk of DS in allele, dominant, recessive, and over-dominant models. RFC1 A80G polymorphism was significantly correlated with having DS risk in both dominant and recessive inheritance comparison models (OR = 2.316, p = 0.03 for the recessive model, and OR = 2.704, p = 0.001 in dominant model). The dominant model is the best inheritance model as it yields the strongest association with the smallest AIC (-8.8). The minor G allele was associated with an increased risk of DS (OR = 2.09, 95% CI = 1.39-3.15, p < 0.001) (Table IV).
3.2. Polymorphism CBS 844in68 and increased risk of having DS baby in mothers
As indicated in the method section, polymorphism CBS 844ins68 was genotyped using PCR and electrophoresis. Figure 3A shows the result of agarose gel electrophoresis for some participants. As expected, a 98 bp fragment was observed for the wild-type homozygote genotype (Ins-/-), while 2 fragments of 98 bp and 166 bp were detected in the heterozygous genotype (Ins+/-). The presence of a 166 bp fragment confirmed the homozygous of (Ins+/Ins+).
The true amplification of the CBS 844ins68 flanking region was confirmed through the sanger sequencing (Figure 3B). The genotype frequency of 844ins68 was compared between DSM and CM (Figure 3C, Table II). As indicated, a significant difference was observed in genotype frequency of CBS 844ins68 between DSM and CM (χ2 = 7.588, p = 0.0225). The observed genotype frequency for the Ins -/- genotype in DSM was 16% compared to 24% in CM (p = 0.206); Ins+/- genotype frequency was 50% in DSM, and 31% in CM which was statistically significant (p = 0.035), and the Ins+/+ genotype was 34% in DSM compared to 45% in CM (p = 0.216). The genotype distribution of CBS 844Ins68 polymorphism significantly differed between CM and DSM (χ2 = 0.112, p < 0.001). The distribution of all CBS genotypes was inconsistent with HWE (χ2 = 12.261, p = 0.737) (Table II, Figure 3C). The allele frequency of 844ins68 is shown in table III. As indicated, the “Ins +” allele frequency was found to be 0.605 in CM and 0.59 in DSM (p = 0.838). The “ins-” allele frequency was 0.395 and 0.41 among CM and DSM, respectively. No significant difference was observed in SNP allelic frequencies of DSM and CM (p = 0.838) (Table III).
3.3. Gene-gene interaction between CBS 844ins68 and RFC1 A80G polymorphisms
The interaction of CBS 844ins68 and RFC1 A80G in all 200 genotyped DSM and CM was summarized in table IV. As illustrated, a combination of both polymorphisms RFC1 A80G and CBS 844ins68 in a homozygous state (GG/Ins+/+) can significantly increase the risk of DS (OR = 6.667 and p = 0.022).







4. Discussion
This is the first study showing the positive association of CBS 844ins68 and RFC1 A80G polymorphism as a risk factor for DS in Iranian young DS mothers who were residents of southwest Iran.
We found that only the heterozygous state of CBS 844ins68 polymorphism was more frequent among DSM than the CMs and half of the DSM were positive for the 844ins variant. The calculated OR was 2.419, which means heterozygous mothers of the 844ins68 variant have around a 2.4-fold increase in the risk of DS than the mothers with other genotypes of 844ins68. The frequency of the 80G allele in DSM (48.5%) is significantly higher than the observed frequency in CM (31%). The OR was calculated as 2.09. We observed that 24% of DSM were homozygous for the 80G variant (GG genotype), and 49% were heterozygous for 80G (AG genotype). In CM, the frequency of GG and AG genotypes were 12% and 38%, respectively.
The gene RFC1 encodes a transporter of folate into and out of cells. Mutation in the RFC1 gene can be associated with multiple defects in mice models of birth defect, and supplementation with folic acid can increase the survival of embryos (9). Through HPA RNA-seq normal tissues, a high expression level of RFC1 was found in the liver, placenta, heart, and whole blood (https://www.ncbi.nlm.nih.gov/gene/6573). RFC1 has not been directly connected to the increased level of total homocysteine, but it may decrease the availability of folic acid by the developing fetus, resulting in growth problems. Different variants of RFC1 can influence the concentration of folate metabolism (10). In the latest release of dbSNP (https://www.ncbi.nlm.nih.gov/snp/), 3027 single nucleotide variants have been reported in the RFC1 gene. However, polymorphism A80G is the most frequent nucleotide change in the RFC1 gene. The A80G is a missense variant resulting in the substitution of histidine amino acid codon (CAG) to arginine (CGG) and altered concentrations of folate metabolic metabolism products (6). Chango et al. reported that the A80G variant of this gene could increase the total homocysteine level in the homozygous GG state (10). Yet, there appears to be little or no consensus about how to the allele inherited and further analysis is needed. However, it is well documented that the transporter protein derived from the RFC1 allele 80G is less functional than the protein encoded by the wild-type allele (80A). It has been documented that women who are homozygous of RFC1 80G have a 2-fold higher risk of having a baby with a birth defect than those with the 80A variant (11). Totally, 6 studies investigated the association of RFC1 A80G polymorphism as a maternal genetic risk factor for DS showing the positive association among Italian, French, Romanian, Turkish, Indian, and Iranian mothers as well (Table V).
Similar to Iranian mothers, the Italian and Indian populations showed a positive association between the presence of RFC1 A80G polymorphism and increased risk of DS. However, no association was observed in a population of Turkish, French, Brazilian, and Romanian. The frequency of the 80G variant in Italian mothers was 49.5%, close to our observed frequency of 48.5% among Iranian mothers (12). This finding was unsurprising as genetic similarity and close relationship between Iranians and Italians based on previously published studies. Iranian-related ancestry was evidenced in Italy's Sardinia and Sicily cities (13, 14). The frequency of the 80G allele was 37.16 among Indian mothers of DS (15). The same story was documented among the Iranian and Indian populations due to the migratory flux between these countries (16). In the case of a population with a lack of association, the unlike results may arise from the difference in their nutritional regime or distinct genetic context.
The CBS, a crucial enzyme in the folic acid cycle, is strongly expressed in normal brain tissues and the liver, pancreas, and kidney. The 844ins68 (Allele ID: 231132) is the most common polymorphism of the CBS gene, which involves a 68-bp insertion in the coding region of exon8 at position 844. It causes a frameshift, introducing 2 nonsense codons in the same exon. The polymorphism duplicates the intron-exon boundary of exon 8, producing an alternative splicing site (17). In 1995, the CBS 844ins68 was first reported in a homocystinuria individuals (18). The current study showed a significant difference in the genotype frequency of CBS 844ins68 between DSM and CM. The “Ins+” allele frequency was low (0.605 in CM and 0.59 in DSM. Extensive ethnic and geographic variability was connected with CBS 844ins68 polymorphism (19, 20) (Table V).
The frequency of the heterozygous genotype of the 844ins68 allele is around 11.7% in normal North American populations; the Netherlands, Spain, the United States, and Ethiopia have an average prevalence of 11-17%, and the population of China, Czechoslovakia, India, Chile, and Colombia have a low prevalence of 1-10% for heterozygous genotype. The 844ins68 allele has not been reported in Indonesia or Japan (21).
Similar to our result, in a study on the frequency of 844ins68 in Southern Iran, this population showed great similarity with other Caucasian populations, especially in South Italy and North America. In contrast, it varied from East Asian and African populations (22). Although the 844ins68 does not seem to disturb the activity of the CBS enzyme, the frequency is increased in people with premature coronary-artery disease (18). The association of CBS 844ins68 with DS has been studied in different populations (Table V), but the results were controversial. Fintelman-Rodrigues et al. in a study on the Brazilian people, showed that CBS 844Ins68 polymorphism did not correlate with DS pregnancies. While the combined genotype of this polymorphism and other polymorphisms of the folic acid cycle, such as RFC1 80G>A, MTRR 66A>G, MTR 2756A>G, MTHFR 1298A>C were associated with the increased risk of having DS babies (23). However, Scala et al. suggested that CBS 844Ins68 could not increase the risk of having a baby with DS in Italy (12). Such contradictory results may relate to various factors, including sample size or differences in geographical area and racial background, which should be considered in future studies. We also found that the combination of both RFC1 GG genotypes with CBS 844ins +/+ can increase the risk of having DS babies to 6.667.
One of the valuable points of this study was the evaluation of RFC1 A80G and CBS 844ins68 in 100 young mothers under 35. It has been shown that most DS children have been born to young women, triggering the idea that other risk factors than increased maternal age may be involved in the etiology of DS (24, 25). In 2009, Migliore and Migheli showed that chromosomal nondisjunction and missegregation are multifactorial traits presented by genetics and environment. They found that young mothers of DS children might have a genetic predisposition (like special polymorphisms) to abnormal chromosome segregation in somatic and germinal cells (26). In this study, we selected mothers with a mean maternal age of 24.03 ± 5.19 yr during the pregnancy of DS babies and excluded those over the age of 35 during the pregnancy. In fact, by considering such criteria, we reduced the effect of maternal age on the incidence of DS. Therefore, in mothers of DS children, the syndrome may have arisen from something other than maternal age. Of note, this idea has not been considered in most of the studies that estimated the association of maternal polymorphisms with having DS babies (15, 27).

5. Conclusion
In conclusion, this study showed that RFC1 A80G and CBS 844Ins68 are 2 polymorphisms of folate metabolism that can be involved in the etiology of DS among Iranian mothers of Khuzestan province.
Data availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author contributions
MTB and NF had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: MTB. Acquisition, analysis, or interpretation of data: MTB, NF, PGH and JMA. Drafting of the manuscript: NF, MTB and PGH. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: NF and MTB. Supervision: MTB and PGH.
Acknowledgments
The authors appreciate the efforts of all colleagues at Ahvaz Jundishapur University of Medical Sciences Research Affairs, Department of Medical Genetics and Cellular and Molecular Research Center for their generous assistance during this project (Grant No: CMRC-9717).
Conflict of Interest
The authors declare that there is no conflict of interest.
Type of Study: Original Article |
Subject:
Reproductive Genetics
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |







