Sat, Apr 26, 2025
[Archive]
Volume 22, Issue 6 (June 2024)
IJRM 2024, 22(6): 481-494 |
Back to browse issues page
Ethics code: IR.ACECR.ROYAN.REC.1395.124 and IR.ACECR.ROYAN.REC.1396.75
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Bagherian E, Jokari S, Borjian Boroujeni P, Haratian K, Sabbaghian M, Mohseni Meybodi A. Role of genetic variations and protein expression of β-Microsemino protein in intrauterine insemination outcome of unexplained infertile men: A case-control study. IJRM 2024; 22 (6) :481-494
URL: http://ijrm.ir/article-1-3212-en.html
URL: http://ijrm.ir/article-1-3212-en.html
Elham Bagherian1 

 , Sahar Jokari1
, Sahar Jokari1 

 , Parnaz Borjian Boroujeni1
, Parnaz Borjian Boroujeni1 

 , Kaveh Haratian2
, Kaveh Haratian2 

 , Marjan Sabbaghian3
, Marjan Sabbaghian3 

 , Anahita Mohseni Meybodi *
, Anahita Mohseni Meybodi * 

 4
4


 , Sahar Jokari1
, Sahar Jokari1 

 , Parnaz Borjian Boroujeni1
, Parnaz Borjian Boroujeni1 

 , Kaveh Haratian2
, Kaveh Haratian2 

 , Marjan Sabbaghian3
, Marjan Sabbaghian3 

 , Anahita Mohseni Meybodi *
, Anahita Mohseni Meybodi * 

 4
4
1- Department of Genetics, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran.
2- Department of Pathology and Laboratory Medicine, Western University, London, Ontario, Canada.
3- Department of Andrology, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran.
4- Department of Genetics, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran. & Department of Pathology and Laboratory Medicine, Western University, London, Ontario, Canada. ,anahitamohseni@gmail.com
2- Department of Pathology and Laboratory Medicine, Western University, London, Ontario, Canada.
3- Department of Andrology, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran.
4- Department of Genetics, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran. & Department of Pathology and Laboratory Medicine, Western University, London, Ontario, Canada. ,
Full-Text [PDF 2293 kb]
(444 Downloads)
| Abstract (HTML) (453 Views)
1. Introduction
According to the World Health Organization (WHO), infertility is defined as the failure to conceive after 12 months or more of regular, unprotected intercourse (1). Unexplained male infertility refers to men who have normal results on standard diagnostic tests but are still unable to impregnate a woman who has no fertility issues herself (2). The prevalence rate of unexplained infertility is 22-30% of all infertile cases (3).
Intrauterine insemination (IUI) is an assisted reproductive method that transfers sperm into a woman’s uterine at the time of ovulation (4). IUI is often used as the first-line treatment for unexplained infertility before considering other invasive and more expensive complex assisted reproductive techniques (ART), such as in vitro fertilization (5, 6).
Several variables can affect the success of IUI, including age and ovarian reserve in females, sperm parameters and semen preparation protocol in males, and type of infertility and its duration (5, 7). A defect in gametic interaction can result in failed fertilization and IUI failure (8). Recent studies have suggested that the sperm proteomic and genomic profile is related to ART success.
β-microsemino protein (MSMB), encoded by the MSMB gene located on chromosome 10 (q11.2) (http://www.genecards.org/), is also known as prostate secretory protein 94. It is an abundant protein in seminal plasma produced by the epithelial cells of the prostate gland. MSMB is a non-glycosylated protein rich in cysteine, localized on the sperm head and neck (9, 10). It has an inhibitory effect on sperm motility and spontaneous acrosome reaction (11), which may affect male fertility (10). MSMB along with the beta-defensin ESP13.2, constitutes 95% of the proteins released from the Macaque sperm surface during capacitation. A recent study confirmed that mutations in beta-defensin 126 gene and its consequent protein expression, have associations with the results of infertility treatment by IUI (10, 12). Another protein, cysteine-rich secretory protein 3 (CRISP-3), is extensively present in human seminal plasma and remains attached to sperm after capacitation and the acrosome reaction (10, 13). CRISP-3 binds to MSMB with very high affinity, forming noncovalent stable complexes (14). Specific amino acids (Y3, F4, P56, and the C-terminal of β-strand) of MSMB, coded by exons 2 and 4, were found to be essential for interacting with CRISP-3. The impact of some polymorphisms in the MSMB 1 region, as well as the impact of protein expression in seminal plasma, has been proven in men with abnormal semen analysis (10).
The present study aims to evaluate the impact of MSMB variations in the promoter and coding sequences of CRISP-3 interacting regions (exon 2 and 4), and the potential impact of its protein expression on IUI success rate in unexplained infertile men for the first time.
2. Materials and Methods
2.1. Participants
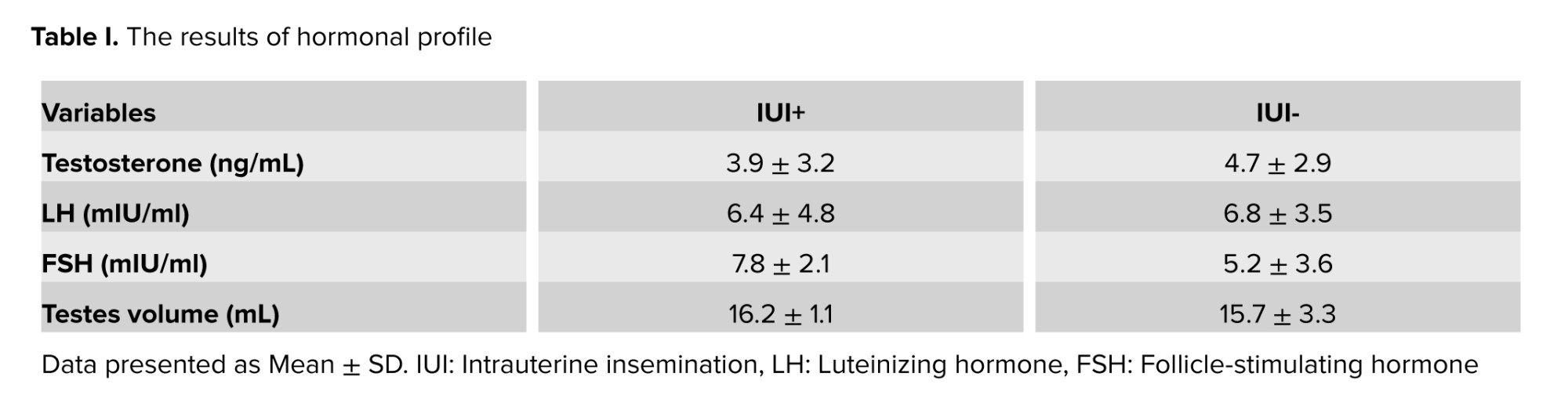
This case-control study involved 100 unexplained infertile men referred to Royan Research Institute from January 2016 to December 2020. The medical histories of all couples were reviewed, and those identified as unexplained infertile couples were selected. Couples with female infertility factors (such as hormonal disorders, irregular menstrual cycles, or abnormalities in the uterus, fallopian tubes, or ovaries) were excluded. The study included 50 men whose wives did not achieve pregnancy after IUI (IUI- group) and 50 men whose wives achieved pregnancy after IUI (IUI+ group). Participants' ages ranged from 20-45 yr (mean age 30.7 ± 6.4 yr), and all had normal semen analysis results.
Sperm count, morphology, and progressive motility were in the normal range according to the WHO 2010 criteria for human semen characteristics (15) (Table I). The study excluded men who had infections of the male accessory glands, hypogonadotropic hypogonadism, clinical varicocele, retractile testis, genital trauma, drug use, concurrent hormonal treatment, Y chromosome microdeletions, or abnormal karyotypes. All men were examined and consulted for any history and/or active genital tract infections that lead to male infertility such as chlamydia, mycoplasma, and mumps. Men with a history of these infections were excluded from the study.
2.2. Sample size
Considering that polymorphism is defined as a DNA variation that occurs in > 1% of the population, the sample size was considered to be 100. Blood samples were taken from 100 men for genotyping tests. Among these participants semen samples were available from 35 participants.
2.3. Polymerase chain reaction (PCR) and sequencing
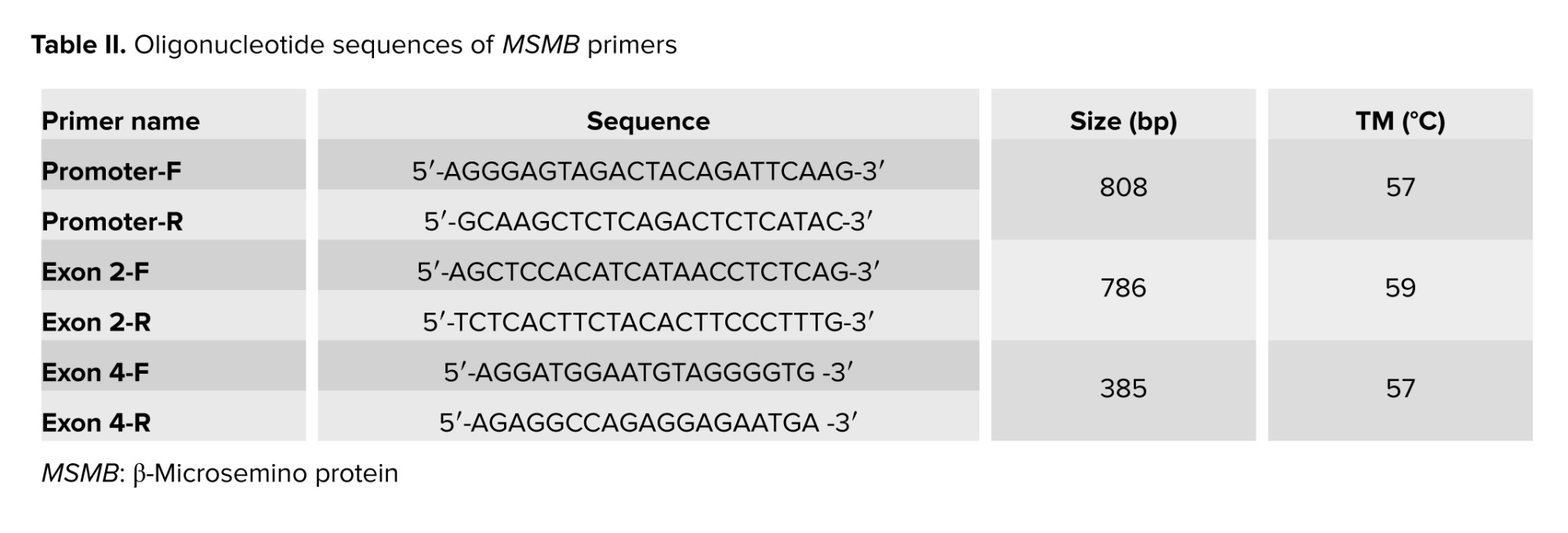
To investigate the genetic variations, total genomic DNA was extracted from peripheral blood utilizing the salting out technique (16). MSMB gene sequence was obtained from the Gene database (http://www.ncbi.nlm.nih.gov). PCR was performed using specific primers designed by Perlprimer v1.1.21 software for promoter region and exon 2 and 4 (Table II).
PCR was performed in a final volume of 30 μl including 50 ng of total DNA, 7 μL of PCR master mix (Ampliqon, Denmark), and 5 μM of each primer. PCR was performed for promotor region using the Eppendorf PCR system as follows: initial denaturation for 4.5 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 1 min, annealing at 59°C for 1 min, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with gel red for analysis.
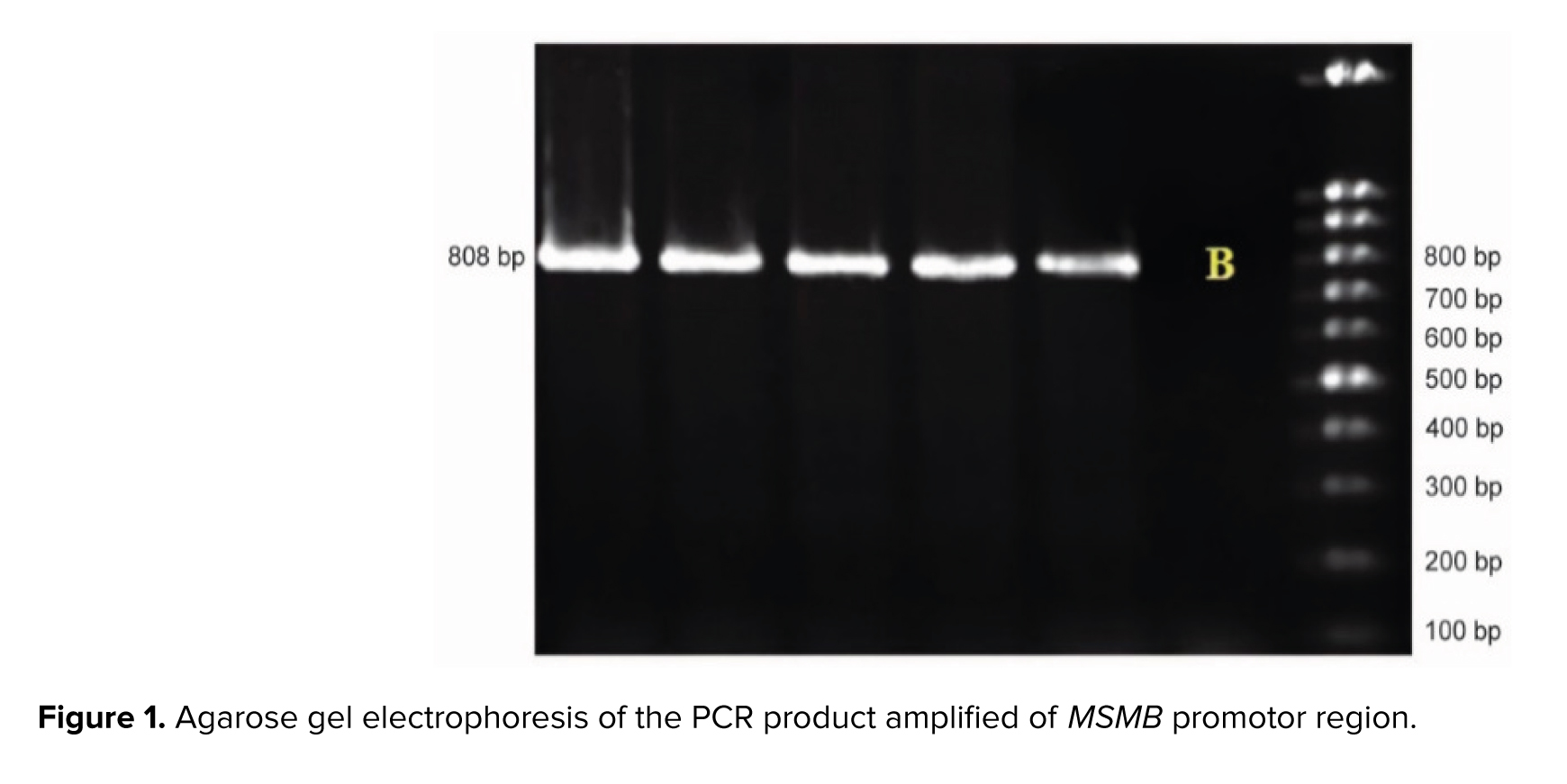
PCR for exon 2 was performed in a final volume of 30 μl including 50 ng of total DNA, 15 μL of PCR Master Mix (Ampliqon, Denmark), 12 μL of dH2O, 1 μM of each primer, and 1 μM of DNA. PCR was done using the Eppendorf PCR system as follows: initial denaturation for 4 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 30 sec, annealing at 59°C for 45 sec, and 1 min at 72°C for extension, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with Gel red for analysis (Figures 1 and 2).
PCR for exon 4 was performed in a final volume of 30 μl including 50 ng of total DNA, 15 μL of PCR Master Mix (Ampliqon, Denmark), 12 μL of dH2O, 1 μM of each primer, and 1 μM of DNA. PCR was performed using the Eppendorf PCR system as follows: initial denaturation for 4 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 30 sec, annealing at 57°C for 1 min and 1 min at 72°C for extension, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with gel red for analysis (Figure 3).
The PCR products were sent to Fazabiotech Co. (Tehran, Iran) for sequencing. Sanger sequencing was conducted using the ABI 3730XL Capillary Sequencer, and the data were then compared with human wildtype MSMB gene sequence at the NCBI-gene database (NC_000010.11 as the reference sequence) to recognize probable variations. FinchTV software v.1.4.0 and align sequences nucleotide BLAST were used to analyze the sequencing results. The genetic variations in the selected regions of MSMB were compared between groups.
2.4. Protein extraction and sandwich enzyme-linked immunosorbent assay (ELISA)
To evaluate sperm protein expression, semen samples were obtained from 35 participants. 20 of them were IUI+ and 15 were IUI-. The samples were collected after 2-5 days of sexual abstinence and stored at -196°C in the liquid nitrogen tank.
Sperm wash was performed using phosphate-buffered saline (PBS) and centrifugation. The frozen semen samples were melted at 0°C. An equal volume of each semen sample and PBS were centrifuged at 1500 rpm for 15 min at 4°C. The supernatant was discarded and 500 μl of PBS was added to the pellet. Centrifugation was performed at 1500 rpm for 10 min at 4°C. The supernatant was discarded and the sperm pellet was stored at -80°C.
Bicinchoninic acid assay was used to determine the total protein concentration. A reduction of Cu+2 to Cu+1 by proteins in an alkaline solution result in the purple-colored reaction product. The amount of this product can be measured by a spectrophotometer to estimate protein concentration. To investigate MSMB expression level ELISA was performed (Abbexa, abx57402). The optical density measurement was performed by absorbance measuring at 450 nm. The absorbance was compared to the standard curve that was obtained using the standard curve of pure MSMB protein.


2.5. Ethical considerations
This study was performed based on the guidelines of the Declaration of Helsinki and was approved by the Ethical Committee of Royan Institute, Tehran, Iran (Code: IR.ACECR.ROYAN.REC.1395.124 and IR.ACECR.ROYAN.REC.1396.75). All participants provided written informed consent.
2.6. Statistical analysis
Statistical analysis was conducted using a statistical package for the social science (SPSS) software (version 20; SPSS, Chicago, IL). The Chi-square test was used to analyze the genetic variables in the studied groups. t test and correlation tests were performed to evaluate the correlation between genetic variations of promoter region and protein expression levels in IUI+ and IUI- groups. ELISA data were also analyzed using the Mann-Whitney U test in men with or without identified Single nucleotide polymorphisms (SNPs) in the promoter region, regardless of their IUI results. Fisher exact test was performed to determine whether the population is in Hardy Weinberg equilibrium. For the Chi-square test, t test, Fisher’s exact test, and Mann-Whitney U test p-value < 0.05 was considered statistically significant, and for the correlation test p-value < 0.01 was considered statistically significant.
3. Results
3.1. Analysis of the MSMB gene alternations
The sequencing analysis among 100 unexplained infertile men revealed 4 nucleotide changes: NC_000010.10:g.51549496T>C (rs10993994T>C) and NC_000010.10:g.51549314C>T (rs12770171C>T) are located in the promoter region, whereas NC_000010.10:g.51556111T>C (rs2075894T>C) and NC_000010.10:g.51556286T>G (rs4517463T>G) are located in the intronic region (RCh37.p13) (Figure 4). Overall, no significant differences were observed between the case and control group (Table III).
The rs10993994T>C variant was detected in heterozygous form in 54% of participants in the IUI+ group and 30% of men in the IUI- group. This SNP was detected in homozygous form in 18% of men in IUI+ and 24% of men in IUI- group. No statistically significant difference was observed between groups (p = 0.624, Table II). The allelic frequency of rs10993994 T>C was 56%: 44% in the IUI+ group and 62%: 38% in the IUI- group and there was no significant difference (p = 0.38). It mediates MSMB prostate secretion levels and is linked to mRNA expression changes. Previous studies have demonstrated that MSMB plays a tumor suppressor role in prostate tissue, and its over-expression can inhibit growth, invasion, and metastasis as well as promote apoptosis in prostate cancer (PCa) cells. The rs10993994 polymorphism was associated with decreased expression of MSMB protein in prostate tissue cells, thus it could be associated with an increased risk of PCa (17, 18) (Table IV).
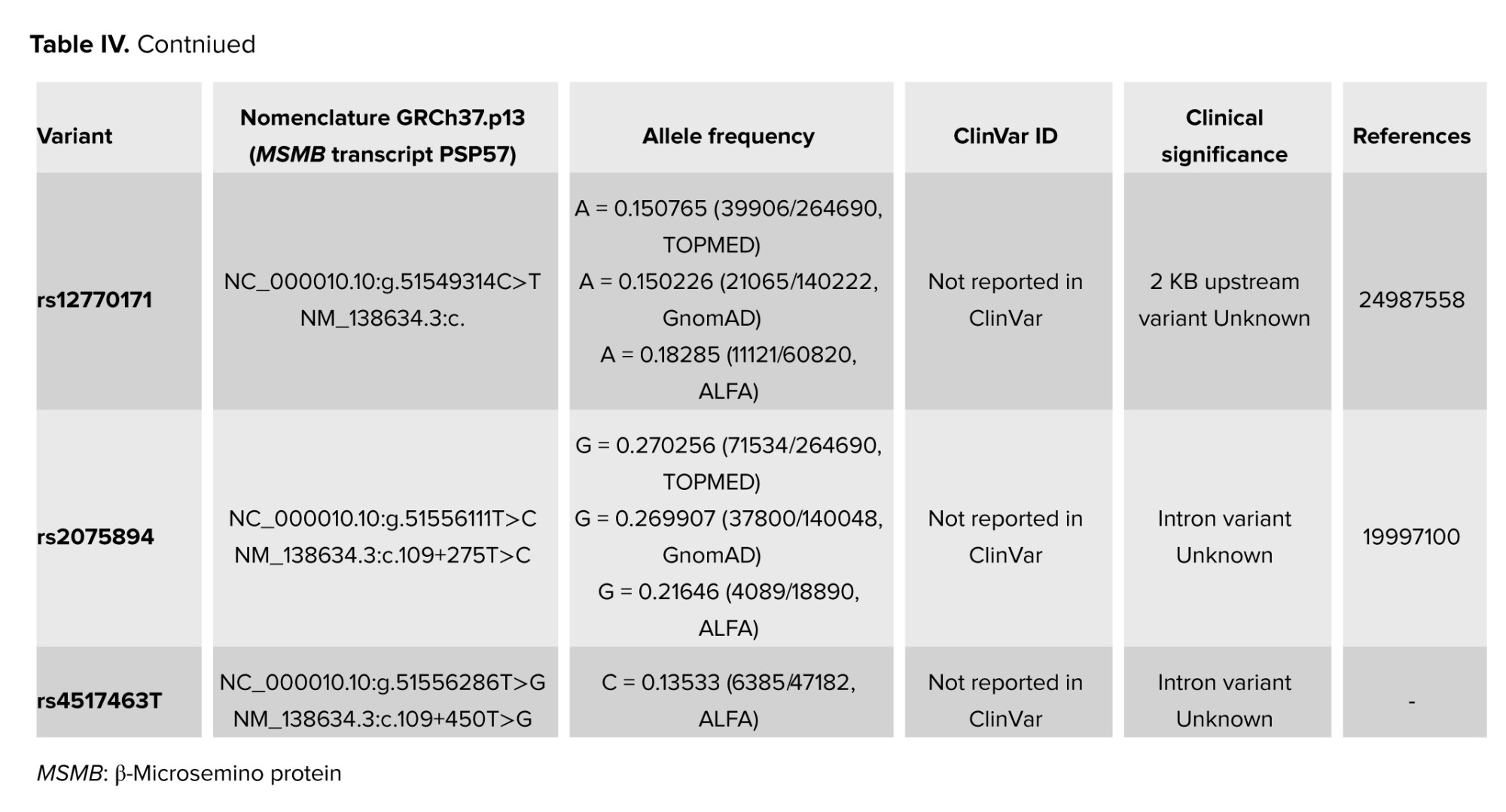
The C>T (rs12770171) variant was detected in heterozygous form in 30% of men in the IUI+ group and 44% of men in the IUI- group. This SNP was detected in homozygous in 4% in the IUI- group and 2% in the IUI+ group. No statistically significant difference was observed between groups (p = 0.25). The allelic frequency of rs12770171 C>T was 16%: 84% in the IUI+ group and 25%: 75% in the IUI- group and there was no significant difference (p = 0.11). This variant has been suggested to be linked with benign prostatic hyperplasia (BPH), the most common prostate disease in aging men as well as PCa in limited studies. This variation has not been previously reported in ClinVar. Meanwhile, based on gnomAD v3.1.2 data, it was identified in 23489/152160 (0.154) of alleles tested from presumed healthy individuals (19, 20) (Table IV).
The distribution of rs2075894T>C was detected in the IUI+ group and the IUI- group. No statistically significant difference was observed between the 2 groups (p = 0.32, Table II). The allelic frequencies of rs2075894T>C in the 2 groups were 80%: 20% in the IUI+ group and 73%: 27% in the IUI- group and there was no significant difference (p = 0.24).
The rs4517463T>G distribution was detected in the IUI+ group and in the IUI- group. No statistically significant difference was observed between IUI+ and IUI- groups (p = 0.40, Table II). The allelic frequencies of rs4517463T>G in the 2 groups were 87%: 13% in the IUI+ group and 86%: 14% in the IUI- group and there was no significant difference (p = 0.48).
There is no record for this variation in ClinVar, besides, it was identified in 39750/151984 (0.261) of alleles tested from presumed healthy individuals in gnomAD v3.1.2. None of the patients who had the abovementioned SNPs in our study showed family history or any evidence of PCa after being examined by the specialist (Table IV).
3.2. Analysis of MSMB expression levels on sperm using ELISA
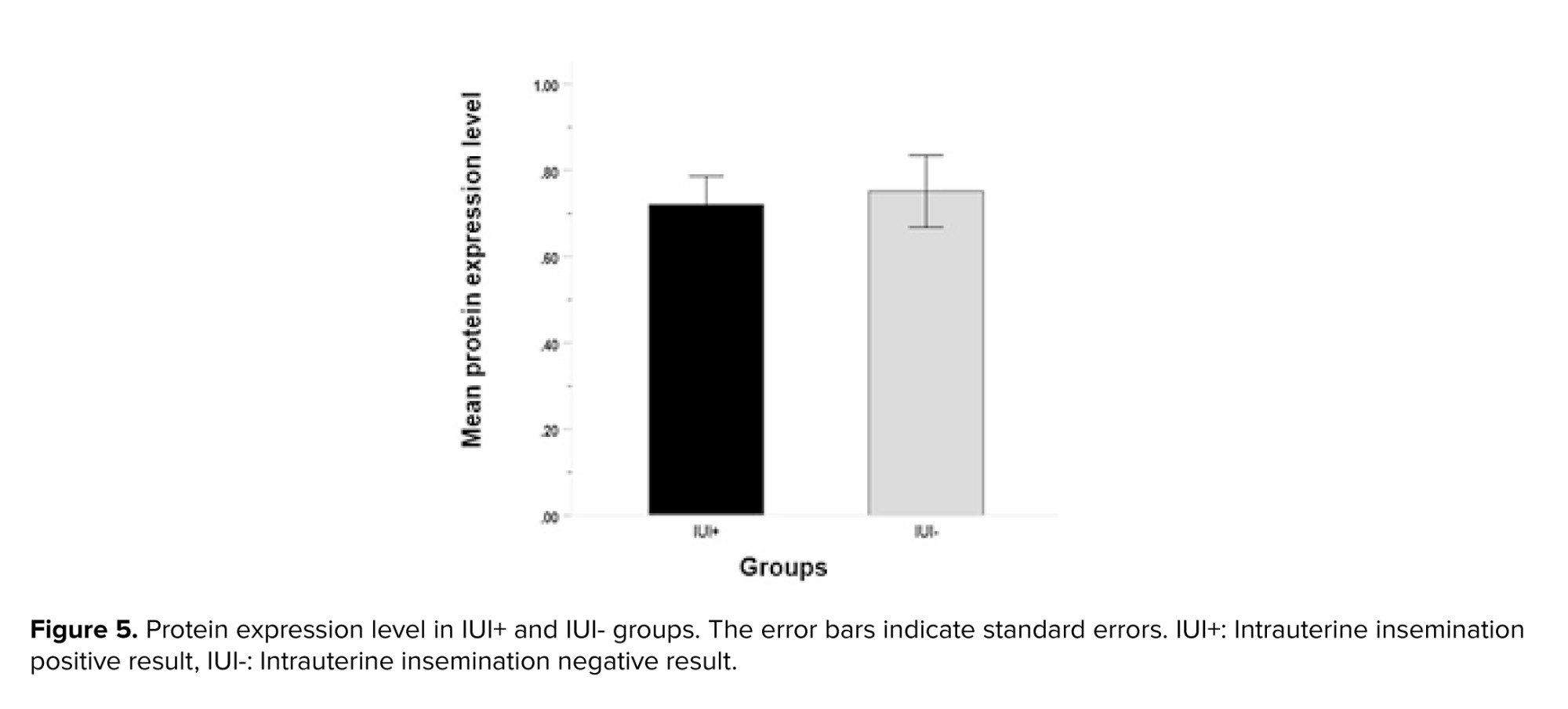
Analysis of protein expression using ELISA revealed no significant differences in MSMB protein values between IUI+ (0.75 ± 0.14 ng/ml) and IUI- groups (0.72 ± 0.13 ng/ml) (p = 0.86, Figure 5).
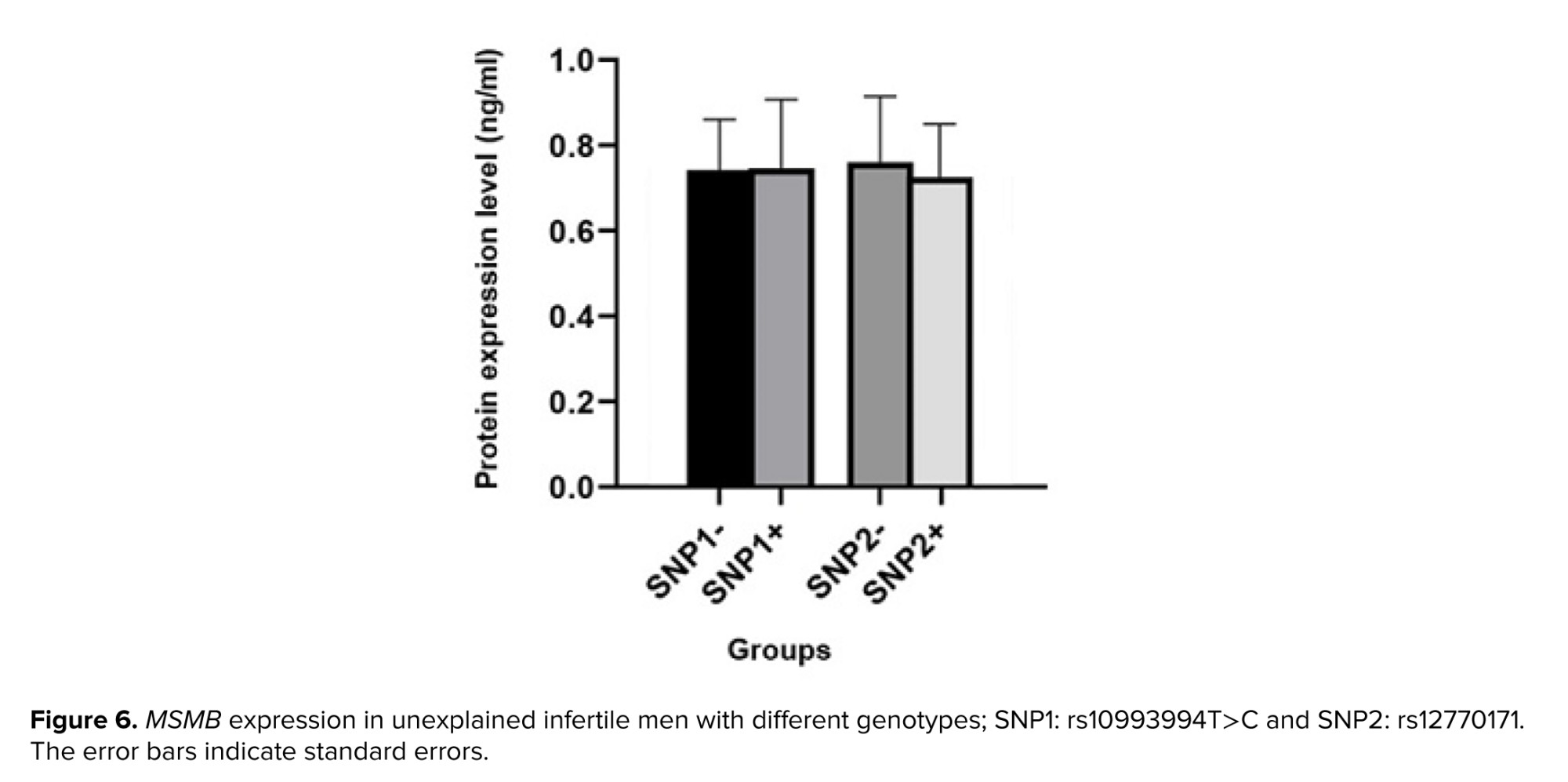
To evaluate the impact of promoter variations on protein expression, semen samples were divided into 2 groups: SNP+ and SNP-. The rs10993994T>C variant (SNP1), was detected in 63 men; however, semen samples from almost half of them were available. Protein expression in semen samples from 17 SNP+ (0.75 ± 0.15 ng/ml) and 14 SNP- (0.74 ± 0.11 ng/ml) were analyzed. Statistical analysis revealed no association between detected genetic variation and protein expression (p = 0.53). The rs12770171 C>T variant (SNP2), was detected in 40 participants. Semen samples were obtained from 14 SNP+ (0.73 ± 0.9 ng/ml) and 17 SNP- (0.73 ± 0.34 ng/ml) (Figure 6). Statistical analysis showed no association between the genetic variation and protein expression (p = 0.13).



4. Discussion
MSMB is a plentiful protein found in seminal plasma and is also detectable on the sperm surface. It is a peripheral protein that binds to membrane integral proteins, which play a role in gamete recognition and sperm-egg interaction at the fertilization site by an inhibitory function on spontaneous acrosome reaction. In addition, with azoospermia, oligoasthenoteratozoospermia, severe teratozoospermia, and asthenozoospermia compared with fertile men (10, 21, 22). Proteomic biomarkers are reliable and invasive means to assess the fertilizing potential of the sperm. Genetic and proteomic markers can be used as predictive tools for ART success, while they cannot be predicted by conventional semen analysis (23, 24). No studies have been found on the genetic variants in the promoter and the coding sequences of interacting domains of the MSMB gene, as well as its expression level in sperm of unexplained infertile men.
So far, many studies confirmed the association of variants detected in the MSMB genes promotor region such as rs10993994T>C (Table V) with PCa risk (21, 22, 25-28). This variation has been previously reported in ClinVar as a risk factor for PCa; however, it was identified in 81774/151992 (0.538) of alleles tested from presumed healthy individuals in (gnomAD v3.1.2). Some studies have demonstrated an association between T allele of rs10993994 in MSMB and increased risk of PCa, while other studies have shown different results about American white and black men and also Japanese men (21). This variant was not found to be a genetic risk factor for BPH. A previous study showed a similar frequency of rs10993994 in BPH and PCa patients and healthy participants (10).
However, most individuals who shared their data with gnomAD did not fully consent to the sharing of detailed clinical information. Consequently, we are usually unable to provide additional details about the phenotype status of variant carriers. Thus, we cannot rule out the possibility that some participants in gnomAD may suffer from infertility. Interestingly, a previous study suggested that subjects with TT genotype had an increased risk of having azoospermia compared with CC genotype. However, no differences in risk were observed for the TT genotype or T allele among men with oligozoospermia. The study suggested that genetic variations on this promoter region could create or demolish some transcription factor binding sites, which may change the MSMB gene expression and consequently impact spermatogenesis (10). The specific mechanism of how the rs10993994 variant contributes to male infertility remains unclear, as there is no direct functional data available for this variation. In our study, the rs10993994 C>T polymorphism was detected in both groups in which the semen analysis of all participants was normal according to WHO 2010 criteria. This variant did not show any significant differences between IUI+ and IUI- groups. Based on a previous study, despite the unclear mechanism of MSMB in PCa, it has been shown that MSMB-derived polypeptide induces prostate cell death. During the process of tumorigenesis the regulatory role of MSMB on cell growth is lost (29). Although this variant was associated with PCa based on some studies, we did not find any PCa symptoms in examining the clinical history of any of participants.
rs12770171 C>T that is located at 242 nucleotides upstream of the transcription initiation site was detected in both groups. This variant did not show any significant differences between the studied groups.
rs2075894T>C and rs4517463T>G are other polymorphisms that were detected downstream of exon 2, but these variants also did not show any significant differences between IUI+ and IUI- groups.
To the best of our knowledge, rs4517463T>G was not mentioned in any publication to be related to any disease, nor been reported in ClinVar. The allele frequency for this variation was 18745/152096 (0.123) in gnomAD v3.1.2. For both intronic variants (rs2075894T>C and rs4517463T>G), their potential impact on splicing was assessed by computational methods for splice site prediction such as SpliceSiteFinder-like, MaxEntScan, NNSPLICE, and GeneSplicer. Based on the data, none of the intronic variants had any significant impact on splicing.
Previous studies have reported that the MSMB protein binds to sites on the surface of human sperm and interacts with an integral protein on the sperm plasma membrane that may act as its receptor (21). In pigs, it has been shown to reduce Na+-K+-ATPase activity and sperm motility. It is believed that sperm hyperactivation plays a vital role in penetration to zona pellucida and successful fertilization. Meanwhile, the MSMB protein may act as an inhibitor of sperm capacitation, thereby preventing premature sperm hyperactivation and premature acrosome reaction before sperm-egg interaction at the fertilization site. These findings collectively suggest that MSMB plays a role in regulating sperm physiology (30).
In another study, the expression of MSMB protein was detected as significantly higher in oligoasthenoteratozoospermic patients’ seminal plasma compared with the fertile group (21). Another published result showed higher expression levels in the seminal plasma of idiopathic infertile males compared with the fertile group (10). Considering that MSMB protein is secreted from the prostate gland and added to semen at the time of ejaculation, genetic variations in this gene apparently cannot affect the spermatogenesis that was previously performed in the testis on the upstream of this pathway, causing azoospermia (30).
Our data for MSMB protein level on sperm samples of unexplained infertile men revealed no significant difference between IUI+ and IUI- groups. Based on the results of previous studies, MSMB level was significantly increased in subfertile (oligozoospermia and azoospermia) cases’ seminal plasma samples when compared with fertile controls (10, 21). The higher level of the protein in the seminal plasma of men with oligozoospermia and azoospermia could be attributed to a diminution in the number of MSMB molecules bound to the spermatozoa due to the absence or low concentration of binding sites for MSMB on the sperm surface which consequently increases the free, unattached protein levels in seminal plasma. In unexplained infertile men, the spermogram was normal, and all the participants in this study showed normal sperm count and motility. Also, we specifically measured the level of protein on the sperm surface and not in seminal plasma, to eliminate the effects of the sperm washing process during the IUI procedure. According to our results, the expression of MSMB protein on sperm could not be a predictor of IUI success rate.

5. Conclusion
In conclusion, our results illustrated that variations detected in the MSMB gene may not be associated with the IUI success rate in unexplained infertile men. The result of the present study also indicates that there is no association between MSMB protein level on sperm surface and IUI success rates in unexplained infertile men. With the limitations of the present study, it should be noted that the sample size was rather small and only included unexplained infertile men. However, further studies using a larger population and including fertile men as a control group can be suggested for these kinds of analysis.
Data availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author contributions
Elham Bagherian, Sahar Jokari, and Parnaz Borjian Broujeni conducted sample preparation, performed tests, analyzed data, and drafted the manuscript. Kaveh Haratian served as the scientific consultant, interpreted data, and revised and edited the paper. Anahita Mohseni Meybodi and Marjan Sabbaghian designed the research study, established inclusion/exclusion criteria, supervised sample management based on these criteria, analyzed and interpreted data, contributed to drafting the manuscript, and revised and edited the paper. All authors reviewed and approved the final version of the manuscript.
Acknowledgments
We express our gratitude to all participants and staff at the Genetic and Andrology Laboratories of Royan Institute, Tehran, Iran, for their technical assistance and services. Artificial intelligence was not utilized in this study.
Conflict of Interest
The authors confirm that they have no conflict of interest to declare.
Full-Text: (69 Views)
1. Introduction
According to the World Health Organization (WHO), infertility is defined as the failure to conceive after 12 months or more of regular, unprotected intercourse (1). Unexplained male infertility refers to men who have normal results on standard diagnostic tests but are still unable to impregnate a woman who has no fertility issues herself (2). The prevalence rate of unexplained infertility is 22-30% of all infertile cases (3).
Intrauterine insemination (IUI) is an assisted reproductive method that transfers sperm into a woman’s uterine at the time of ovulation (4). IUI is often used as the first-line treatment for unexplained infertility before considering other invasive and more expensive complex assisted reproductive techniques (ART), such as in vitro fertilization (5, 6).
Several variables can affect the success of IUI, including age and ovarian reserve in females, sperm parameters and semen preparation protocol in males, and type of infertility and its duration (5, 7). A defect in gametic interaction can result in failed fertilization and IUI failure (8). Recent studies have suggested that the sperm proteomic and genomic profile is related to ART success.
β-microsemino protein (MSMB), encoded by the MSMB gene located on chromosome 10 (q11.2) (http://www.genecards.org/), is also known as prostate secretory protein 94. It is an abundant protein in seminal plasma produced by the epithelial cells of the prostate gland. MSMB is a non-glycosylated protein rich in cysteine, localized on the sperm head and neck (9, 10). It has an inhibitory effect on sperm motility and spontaneous acrosome reaction (11), which may affect male fertility (10). MSMB along with the beta-defensin ESP13.2, constitutes 95% of the proteins released from the Macaque sperm surface during capacitation. A recent study confirmed that mutations in beta-defensin 126 gene and its consequent protein expression, have associations with the results of infertility treatment by IUI (10, 12). Another protein, cysteine-rich secretory protein 3 (CRISP-3), is extensively present in human seminal plasma and remains attached to sperm after capacitation and the acrosome reaction (10, 13). CRISP-3 binds to MSMB with very high affinity, forming noncovalent stable complexes (14). Specific amino acids (Y3, F4, P56, and the C-terminal of β-strand) of MSMB, coded by exons 2 and 4, were found to be essential for interacting with CRISP-3. The impact of some polymorphisms in the MSMB 1 region, as well as the impact of protein expression in seminal plasma, has been proven in men with abnormal semen analysis (10).
The present study aims to evaluate the impact of MSMB variations in the promoter and coding sequences of CRISP-3 interacting regions (exon 2 and 4), and the potential impact of its protein expression on IUI success rate in unexplained infertile men for the first time.
2. Materials and Methods
2.1. Participants
This case-control study involved 100 unexplained infertile men referred to Royan Research Institute from January 2016 to December 2020. The medical histories of all couples were reviewed, and those identified as unexplained infertile couples were selected. Couples with female infertility factors (such as hormonal disorders, irregular menstrual cycles, or abnormalities in the uterus, fallopian tubes, or ovaries) were excluded. The study included 50 men whose wives did not achieve pregnancy after IUI (IUI- group) and 50 men whose wives achieved pregnancy after IUI (IUI+ group). Participants' ages ranged from 20-45 yr (mean age 30.7 ± 6.4 yr), and all had normal semen analysis results.
Sperm count, morphology, and progressive motility were in the normal range according to the WHO 2010 criteria for human semen characteristics (15) (Table I). The study excluded men who had infections of the male accessory glands, hypogonadotropic hypogonadism, clinical varicocele, retractile testis, genital trauma, drug use, concurrent hormonal treatment, Y chromosome microdeletions, or abnormal karyotypes. All men were examined and consulted for any history and/or active genital tract infections that lead to male infertility such as chlamydia, mycoplasma, and mumps. Men with a history of these infections were excluded from the study.
2.2. Sample size
Considering that polymorphism is defined as a DNA variation that occurs in > 1% of the population, the sample size was considered to be 100. Blood samples were taken from 100 men for genotyping tests. Among these participants semen samples were available from 35 participants.
2.3. Polymerase chain reaction (PCR) and sequencing
To investigate the genetic variations, total genomic DNA was extracted from peripheral blood utilizing the salting out technique (16). MSMB gene sequence was obtained from the Gene database (http://www.ncbi.nlm.nih.gov). PCR was performed using specific primers designed by Perlprimer v1.1.21 software for promoter region and exon 2 and 4 (Table II).
PCR was performed in a final volume of 30 μl including 50 ng of total DNA, 7 μL of PCR master mix (Ampliqon, Denmark), and 5 μM of each primer. PCR was performed for promotor region using the Eppendorf PCR system as follows: initial denaturation for 4.5 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 1 min, annealing at 59°C for 1 min, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with gel red for analysis.
PCR for exon 2 was performed in a final volume of 30 μl including 50 ng of total DNA, 15 μL of PCR Master Mix (Ampliqon, Denmark), 12 μL of dH2O, 1 μM of each primer, and 1 μM of DNA. PCR was done using the Eppendorf PCR system as follows: initial denaturation for 4 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 30 sec, annealing at 59°C for 45 sec, and 1 min at 72°C for extension, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with Gel red for analysis (Figures 1 and 2).
PCR for exon 4 was performed in a final volume of 30 μl including 50 ng of total DNA, 15 μL of PCR Master Mix (Ampliqon, Denmark), 12 μL of dH2O, 1 μM of each primer, and 1 μM of DNA. PCR was performed using the Eppendorf PCR system as follows: initial denaturation for 4 min at 94°C, amplification for 35 cycles consisting of denaturation at 94°C for 30 sec, annealing at 57°C for 1 min and 1 min at 72°C for extension, followed by a final extension of 8 min at 72°C. PCR products were run on 1.2% agarose gel and stained with gel red for analysis (Figure 3).
The PCR products were sent to Fazabiotech Co. (Tehran, Iran) for sequencing. Sanger sequencing was conducted using the ABI 3730XL Capillary Sequencer, and the data were then compared with human wildtype MSMB gene sequence at the NCBI-gene database (NC_000010.11 as the reference sequence) to recognize probable variations. FinchTV software v.1.4.0 and align sequences nucleotide BLAST were used to analyze the sequencing results. The genetic variations in the selected regions of MSMB were compared between groups.
2.4. Protein extraction and sandwich enzyme-linked immunosorbent assay (ELISA)
To evaluate sperm protein expression, semen samples were obtained from 35 participants. 20 of them were IUI+ and 15 were IUI-. The samples were collected after 2-5 days of sexual abstinence and stored at -196°C in the liquid nitrogen tank.
Sperm wash was performed using phosphate-buffered saline (PBS) and centrifugation. The frozen semen samples were melted at 0°C. An equal volume of each semen sample and PBS were centrifuged at 1500 rpm for 15 min at 4°C. The supernatant was discarded and 500 μl of PBS was added to the pellet. Centrifugation was performed at 1500 rpm for 10 min at 4°C. The supernatant was discarded and the sperm pellet was stored at -80°C.
Bicinchoninic acid assay was used to determine the total protein concentration. A reduction of Cu+2 to Cu+1 by proteins in an alkaline solution result in the purple-colored reaction product. The amount of this product can be measured by a spectrophotometer to estimate protein concentration. To investigate MSMB expression level ELISA was performed (Abbexa, abx57402). The optical density measurement was performed by absorbance measuring at 450 nm. The absorbance was compared to the standard curve that was obtained using the standard curve of pure MSMB protein.





2.5. Ethical considerations
This study was performed based on the guidelines of the Declaration of Helsinki and was approved by the Ethical Committee of Royan Institute, Tehran, Iran (Code: IR.ACECR.ROYAN.REC.1395.124 and IR.ACECR.ROYAN.REC.1396.75). All participants provided written informed consent.
2.6. Statistical analysis
Statistical analysis was conducted using a statistical package for the social science (SPSS) software (version 20; SPSS, Chicago, IL). The Chi-square test was used to analyze the genetic variables in the studied groups. t test and correlation tests were performed to evaluate the correlation between genetic variations of promoter region and protein expression levels in IUI+ and IUI- groups. ELISA data were also analyzed using the Mann-Whitney U test in men with or without identified Single nucleotide polymorphisms (SNPs) in the promoter region, regardless of their IUI results. Fisher exact test was performed to determine whether the population is in Hardy Weinberg equilibrium. For the Chi-square test, t test, Fisher’s exact test, and Mann-Whitney U test p-value < 0.05 was considered statistically significant, and for the correlation test p-value < 0.01 was considered statistically significant.
3. Results
3.1. Analysis of the MSMB gene alternations
The sequencing analysis among 100 unexplained infertile men revealed 4 nucleotide changes: NC_000010.10:g.51549496T>C (rs10993994T>C) and NC_000010.10:g.51549314C>T (rs12770171C>T) are located in the promoter region, whereas NC_000010.10:g.51556111T>C (rs2075894T>C) and NC_000010.10:g.51556286T>G (rs4517463T>G) are located in the intronic region (RCh37.p13) (Figure 4). Overall, no significant differences were observed between the case and control group (Table III).
The rs10993994T>C variant was detected in heterozygous form in 54% of participants in the IUI+ group and 30% of men in the IUI- group. This SNP was detected in homozygous form in 18% of men in IUI+ and 24% of men in IUI- group. No statistically significant difference was observed between groups (p = 0.624, Table II). The allelic frequency of rs10993994 T>C was 56%: 44% in the IUI+ group and 62%: 38% in the IUI- group and there was no significant difference (p = 0.38). It mediates MSMB prostate secretion levels and is linked to mRNA expression changes. Previous studies have demonstrated that MSMB plays a tumor suppressor role in prostate tissue, and its over-expression can inhibit growth, invasion, and metastasis as well as promote apoptosis in prostate cancer (PCa) cells. The rs10993994 polymorphism was associated with decreased expression of MSMB protein in prostate tissue cells, thus it could be associated with an increased risk of PCa (17, 18) (Table IV).
The C>T (rs12770171) variant was detected in heterozygous form in 30% of men in the IUI+ group and 44% of men in the IUI- group. This SNP was detected in homozygous in 4% in the IUI- group and 2% in the IUI+ group. No statistically significant difference was observed between groups (p = 0.25). The allelic frequency of rs12770171 C>T was 16%: 84% in the IUI+ group and 25%: 75% in the IUI- group and there was no significant difference (p = 0.11). This variant has been suggested to be linked with benign prostatic hyperplasia (BPH), the most common prostate disease in aging men as well as PCa in limited studies. This variation has not been previously reported in ClinVar. Meanwhile, based on gnomAD v3.1.2 data, it was identified in 23489/152160 (0.154) of alleles tested from presumed healthy individuals (19, 20) (Table IV).
The distribution of rs2075894T>C was detected in the IUI+ group and the IUI- group. No statistically significant difference was observed between the 2 groups (p = 0.32, Table II). The allelic frequencies of rs2075894T>C in the 2 groups were 80%: 20% in the IUI+ group and 73%: 27% in the IUI- group and there was no significant difference (p = 0.24).
The rs4517463T>G distribution was detected in the IUI+ group and in the IUI- group. No statistically significant difference was observed between IUI+ and IUI- groups (p = 0.40, Table II). The allelic frequencies of rs4517463T>G in the 2 groups were 87%: 13% in the IUI+ group and 86%: 14% in the IUI- group and there was no significant difference (p = 0.48).
There is no record for this variation in ClinVar, besides, it was identified in 39750/151984 (0.261) of alleles tested from presumed healthy individuals in gnomAD v3.1.2. None of the patients who had the abovementioned SNPs in our study showed family history or any evidence of PCa after being examined by the specialist (Table IV).
3.2. Analysis of MSMB expression levels on sperm using ELISA
Analysis of protein expression using ELISA revealed no significant differences in MSMB protein values between IUI+ (0.75 ± 0.14 ng/ml) and IUI- groups (0.72 ± 0.13 ng/ml) (p = 0.86, Figure 5).
To evaluate the impact of promoter variations on protein expression, semen samples were divided into 2 groups: SNP+ and SNP-. The rs10993994T>C variant (SNP1), was detected in 63 men; however, semen samples from almost half of them were available. Protein expression in semen samples from 17 SNP+ (0.75 ± 0.15 ng/ml) and 14 SNP- (0.74 ± 0.11 ng/ml) were analyzed. Statistical analysis revealed no association between detected genetic variation and protein expression (p = 0.53). The rs12770171 C>T variant (SNP2), was detected in 40 participants. Semen samples were obtained from 14 SNP+ (0.73 ± 0.9 ng/ml) and 17 SNP- (0.73 ± 0.34 ng/ml) (Figure 6). Statistical analysis showed no association between the genetic variation and protein expression (p = 0.13).






4. Discussion
MSMB is a plentiful protein found in seminal plasma and is also detectable on the sperm surface. It is a peripheral protein that binds to membrane integral proteins, which play a role in gamete recognition and sperm-egg interaction at the fertilization site by an inhibitory function on spontaneous acrosome reaction. In addition, with azoospermia, oligoasthenoteratozoospermia, severe teratozoospermia, and asthenozoospermia compared with fertile men (10, 21, 22). Proteomic biomarkers are reliable and invasive means to assess the fertilizing potential of the sperm. Genetic and proteomic markers can be used as predictive tools for ART success, while they cannot be predicted by conventional semen analysis (23, 24). No studies have been found on the genetic variants in the promoter and the coding sequences of interacting domains of the MSMB gene, as well as its expression level in sperm of unexplained infertile men.
So far, many studies confirmed the association of variants detected in the MSMB genes promotor region such as rs10993994T>C (Table V) with PCa risk (21, 22, 25-28). This variation has been previously reported in ClinVar as a risk factor for PCa; however, it was identified in 81774/151992 (0.538) of alleles tested from presumed healthy individuals in (gnomAD v3.1.2). Some studies have demonstrated an association between T allele of rs10993994 in MSMB and increased risk of PCa, while other studies have shown different results about American white and black men and also Japanese men (21). This variant was not found to be a genetic risk factor for BPH. A previous study showed a similar frequency of rs10993994 in BPH and PCa patients and healthy participants (10).
However, most individuals who shared their data with gnomAD did not fully consent to the sharing of detailed clinical information. Consequently, we are usually unable to provide additional details about the phenotype status of variant carriers. Thus, we cannot rule out the possibility that some participants in gnomAD may suffer from infertility. Interestingly, a previous study suggested that subjects with TT genotype had an increased risk of having azoospermia compared with CC genotype. However, no differences in risk were observed for the TT genotype or T allele among men with oligozoospermia. The study suggested that genetic variations on this promoter region could create or demolish some transcription factor binding sites, which may change the MSMB gene expression and consequently impact spermatogenesis (10). The specific mechanism of how the rs10993994 variant contributes to male infertility remains unclear, as there is no direct functional data available for this variation. In our study, the rs10993994 C>T polymorphism was detected in both groups in which the semen analysis of all participants was normal according to WHO 2010 criteria. This variant did not show any significant differences between IUI+ and IUI- groups. Based on a previous study, despite the unclear mechanism of MSMB in PCa, it has been shown that MSMB-derived polypeptide induces prostate cell death. During the process of tumorigenesis the regulatory role of MSMB on cell growth is lost (29). Although this variant was associated with PCa based on some studies, we did not find any PCa symptoms in examining the clinical history of any of participants.
rs12770171 C>T that is located at 242 nucleotides upstream of the transcription initiation site was detected in both groups. This variant did not show any significant differences between the studied groups.
rs2075894T>C and rs4517463T>G are other polymorphisms that were detected downstream of exon 2, but these variants also did not show any significant differences between IUI+ and IUI- groups.
To the best of our knowledge, rs4517463T>G was not mentioned in any publication to be related to any disease, nor been reported in ClinVar. The allele frequency for this variation was 18745/152096 (0.123) in gnomAD v3.1.2. For both intronic variants (rs2075894T>C and rs4517463T>G), their potential impact on splicing was assessed by computational methods for splice site prediction such as SpliceSiteFinder-like, MaxEntScan, NNSPLICE, and GeneSplicer. Based on the data, none of the intronic variants had any significant impact on splicing.
Previous studies have reported that the MSMB protein binds to sites on the surface of human sperm and interacts with an integral protein on the sperm plasma membrane that may act as its receptor (21). In pigs, it has been shown to reduce Na+-K+-ATPase activity and sperm motility. It is believed that sperm hyperactivation plays a vital role in penetration to zona pellucida and successful fertilization. Meanwhile, the MSMB protein may act as an inhibitor of sperm capacitation, thereby preventing premature sperm hyperactivation and premature acrosome reaction before sperm-egg interaction at the fertilization site. These findings collectively suggest that MSMB plays a role in regulating sperm physiology (30).
In another study, the expression of MSMB protein was detected as significantly higher in oligoasthenoteratozoospermic patients’ seminal plasma compared with the fertile group (21). Another published result showed higher expression levels in the seminal plasma of idiopathic infertile males compared with the fertile group (10). Considering that MSMB protein is secreted from the prostate gland and added to semen at the time of ejaculation, genetic variations in this gene apparently cannot affect the spermatogenesis that was previously performed in the testis on the upstream of this pathway, causing azoospermia (30).
Our data for MSMB protein level on sperm samples of unexplained infertile men revealed no significant difference between IUI+ and IUI- groups. Based on the results of previous studies, MSMB level was significantly increased in subfertile (oligozoospermia and azoospermia) cases’ seminal plasma samples when compared with fertile controls (10, 21). The higher level of the protein in the seminal plasma of men with oligozoospermia and azoospermia could be attributed to a diminution in the number of MSMB molecules bound to the spermatozoa due to the absence or low concentration of binding sites for MSMB on the sperm surface which consequently increases the free, unattached protein levels in seminal plasma. In unexplained infertile men, the spermogram was normal, and all the participants in this study showed normal sperm count and motility. Also, we specifically measured the level of protein on the sperm surface and not in seminal plasma, to eliminate the effects of the sperm washing process during the IUI procedure. According to our results, the expression of MSMB protein on sperm could not be a predictor of IUI success rate.

5. Conclusion
In conclusion, our results illustrated that variations detected in the MSMB gene may not be associated with the IUI success rate in unexplained infertile men. The result of the present study also indicates that there is no association between MSMB protein level on sperm surface and IUI success rates in unexplained infertile men. With the limitations of the present study, it should be noted that the sample size was rather small and only included unexplained infertile men. However, further studies using a larger population and including fertile men as a control group can be suggested for these kinds of analysis.
Data availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author contributions
Elham Bagherian, Sahar Jokari, and Parnaz Borjian Broujeni conducted sample preparation, performed tests, analyzed data, and drafted the manuscript. Kaveh Haratian served as the scientific consultant, interpreted data, and revised and edited the paper. Anahita Mohseni Meybodi and Marjan Sabbaghian designed the research study, established inclusion/exclusion criteria, supervised sample management based on these criteria, analyzed and interpreted data, contributed to drafting the manuscript, and revised and edited the paper. All authors reviewed and approved the final version of the manuscript.
Acknowledgments
We express our gratitude to all participants and staff at the Genetic and Andrology Laboratories of Royan Institute, Tehran, Iran, for their technical assistance and services. Artificial intelligence was not utilized in this study.
Conflict of Interest
The authors confirm that they have no conflict of interest to declare.
Type of Study: Original Article |
Subject:
Reproductive Genetics
References
1. Dugule DS-G, Gana ED, Paul J. Causes, effects and management of infertility among reproductive women in Karim Lamido Local Government Area of Taraba State-Nigeria. Texas J Med Sci 2023; 16: 36-49. [DOI:10.62480/tjms.2023.vol16.pp36-49]
2. Panner Selvam MK, Agarwal A, Pushparaj PN, Baskaran S, Bendou H. Sperm proteome analysis and identification of fertility-associated biomarkers in unexplained male infertility. Genes 2019; 10: 522. [DOI:10.3390/genes10070522]
3. Kamath MS, Rikken JF, Bosteels J. Does laparoscopy and hysteroscopy have a place in the diagnosis of unexplained infertility? Semin Reprod Med 2020; 38: 29-35. [DOI:10.1055/s-0040-1718942]
4. Kawale SP, Ambad R, Jha RK, Jha RK, Gaikwad A. Role of artificial intelligence in ICSI (intra-cytoplasmic sperm injection) & IUI (intra-uterine insemination). E3S Web of Conferences; 2024 January 11-12: India. [DOI:10.1051/e3sconf/202449103019]
5. Erdem M, Erdem A, Firat Mutlu M, Ozisik S, Yildiz S, Guler I, et al. The impact of sperm morphology on the outcome of intrauterine insemination cycles with gonadotropins in unexplained and male subfertility. Eur J Obstet Gynecol Reprod Biol 2016; 197: 120-124. [DOI:10.1016/j.ejogrb.2015.12.014]
6. Aboutorabi R, Zamani S, Zarrin Y, Mostafavi FS. A survey on main semen parameters in natural pregnancy and intrauterine insemination: Are there any significant differences? Am J Mens Health 2018; 12: 617-623. [DOI:10.1177/1557988316647966]
7. Atasever M, Kalem MN, Hatırnaz Ş, Hatırnaz E, Kalem Z, Kalaylıoğlu Z. Factors affecting clinical pregnancy rates after IUI for the treatment of unexplained infertility and mild male subfertility. J Turk Ger Gynecol Assoc 2016; 17: 134-138. [DOI:10.5152/jtgga.2016.16056]
8. Siu KK, Serrão VHB, Ziyyat A, Lee JE. The cell biology of fertilization: Gamete attachment and fusion. J Cell Biol 2021; 220: e202102146. [DOI:10.1083/jcb.202102146]
9. Tumova L, Zigo M, Sutovsky P, Sedmikova M, Postlerova P. The ubiquitin-proteasome system does not regulate the degradation of porcine β-microseminoprotein during sperm capacitation. Int J Mol Sci 2020; 21: 4151. [DOI:10.3390/ijms21114151]
10. Anklesaria JH, Mhatre DR, Mahale SD. Structural and molecular biology of PSP94: Its significance in prostate pathophysiology. Front Biosci 2018; 23: 535-562. [DOI:10.2741/4604]
11. Grande G, Vincenzoni F, Mancini F, Barrachina F, Giampietro A, Castagnola M, et al. Quantitative analysis of the seminal plasma proteome in secondary hypogonadism. J Clin Med 2019; 8: 2128. [DOI:10.3390/jcm8122128]
12. Rostami Chayjan M, Sabbaghian M, Alikhani M, Sahraneshin Samani F, Salman Yazdi R, Almadani SN, et al. [Relation of β-defensin 126 gene (DEFB126) alteration and protein expression with unexplained male infertility in Iranian population]. J Arak Univ Med Sci 2014; 17: 31-40. (in Persian)
13. Bu Y, Wang P, Li S, Li L, Zhang S, Wei H. Semen protein CRISP3 promotes reproductive performance of boars through immunomodulation. Int J Mol Sci 2024; 25: 2264. [DOI:10.3390/ijms25042264]
14. El Atab O, Kocabey AE, Asojo OA, Schneiter R. Prostate secretory protein 94 inhibits sterol binding and export by the mammalian CAP protein CRISP2 in a calcium-sensitive manner. J Biol Chem 2022; 298: 101600. [DOI:10.1016/j.jbc.2022.101600]
15. World Health Organization. WHO laboratory manual for the examination and processing of human semen. Switzerland: World Health Organization Press; 2021.
16. Dairawan M, Shetty PJ. The evolution of DNA extraction methods. Am J Biomed Sci Res 2020; 8: 39-45. [DOI:10.34297/AJBSR.2020.08.001234]
17. Mendivelso González DF, Sánchez Villalobos SA, Ramos AE, Montero Ovalle WJ, Serrano López ML. Single nucleotide polymorphisms associated with prostate cancer progression: A systematic review. Cancer Invest 2024; 42: 75-96. [DOI:10.1080/07357907.2023.2291776]
18. Dariane C, Clairefond S, Péant B, Communal L, Thian Z, Ouellet V, et al. High keratin-7 expression in benign peri-tumoral prostatic glands is predictive of bone metastasis onset and prostate cancer-specific mortality. Cancers 2022; 14: 1623. [DOI:10.3390/cancers14071623]
19. Nasimi H, Madsen JS, Zedan AH, Malmendal A, Osther PJS, Alatraktchi FAa. Protein biomarker detection in prostate cancer: A comprehensive review of electrochemical biosensors. Sensors and Actuators Reports 2023; 6: 100168. [DOI:10.1016/j.snr.2023.100168]
20. Carvalho AdL, Pinto SA, Santos WGd. CRISP3 glycoprotein: A good biomarker for prostate cancer? J Brasil Patol Med Lab 2021; 57: e3222021. [DOI:10.5935/1676-2444.20210012]
21. Fu S, Huang Y-L, Luan T, Li N, Wang H-F, Wang J-S. A meta-analysis of influence of MSMB promoter rs10993994 polymorphisms on prostate cancer risk. Eur Rev Med Pharmacol Sci 2019; 23: 9295-9303.
22. Tian P, Zhong M, Wei G-H. Mechanistic insights into genetic susceptibility to prostate cancer. Cancer Lett 2021; 522: 155-163. [DOI:10.1016/j.canlet.2021.09.025]
23. Frapsauce C, Pionneau C, Bouley J, Delarouziere V, Berthaut I, Ravel C, et al. Proteomic identification of target proteins in normal but nonfertilizing sperm. Fertil Steril 2014; 102: 372-380. [DOI:10.1016/j.fertnstert.2014.04.039]
24. Agarwal A, Selvam MKP, Baskaran S. Proteomic analyses of human sperm cells: Understanding the role of proteins and molecular pathways affecting male reproductive health. Int J Mol Sci 2020; 21: 1621. [DOI:10.3390/ijms21051621]
25. Emami NC, Kachuri L, Meyers TJ, Das R, Hoffman JD, Hoffmann TJ, et al. Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nat Commun 2019; 10: 3107.
https://doi.org/10.1038/s41467-019-11810-9 [DOI:10.1038/s41467-019-10808-7]
26. Trujillo-Cáceres SJ, Torres-Sánchez L, Burguete-García AI, Orihuela YCO, Vázquez-Salas RA, Álvarez-Topete E, et al. Contribution of MSMB promoter region gene polymorphism to early-onset prostate cancer risk in Mexican males. Oncotarget 2019; 10: 738. [DOI:10.18632/oncotarget.26592]
27. Labani M, Beheshti A, Argha A, Alinejad-Rokny H. A Comprehensive investigation of genomic variants in prostate cancer reveals 30 putative regulatory variants. Int J Mol Sci 2023; 24: 2472. [DOI:10.3390/ijms24032472]
28. Samtal Ch, El Jaddaoui I, Hamdi S, Bouguenouch L, Ouldim K, Nejjari C, et al. Review of prostate cancer genomic studies in Africa. Front Genet 2022; 13: 911101. [DOI:10.3389/fgene.2022.911101]
29. Desai TA, Hedman AK, Dimitriou M, Koprulu M, Figiel S, Yin W, et al. Identifying proteomic risk factors for overall, aggressive and early onset prostate cancer using Mendelian randomization and tumor spatial transcriptomics. medRxiv 2023. (preprint) [DOI:10.1101/2023.09.21.23295864]
30. Jodar M, Soler-Ventura A, Oliva R, Molecular Biology of Reproduction and Development Research Group. Semen proteomics and male infertility. J Proteomics 2017; 162: 125-134. [DOI:10.1016/j.jprot.2016.08.018]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |




