Sun, Jul 13, 2025
[Archive]
Volume 19, Issue 3 (March 2021)
IJRM 2021, 19(3): 217-226 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Argyraki M, Katafigiotis S, Vavilis T, Papadopoulou Z, Tzimagiorgis G, Haidich A, et al . Influence of conception and delivery mode on stress response marker Oct4B1 and imprinted gene expression related to embryo development: A cohort study. IJRM 2021; 19 (3) :217-226
URL: http://ijrm.ir/article-1-1483-en.html
URL: http://ijrm.ir/article-1-1483-en.html
Maria Argyraki *1 

 , Socrates Katafigiotis2
, Socrates Katafigiotis2 

 , Theofanis Vavilis3
, Theofanis Vavilis3 

 , Zoe Papadopoulou4
, Zoe Papadopoulou4 

 , Giorgos Tzimagiorgis5
, Giorgos Tzimagiorgis5 

 , Anna-Bettina Haidich6
, Anna-Bettina Haidich6 

 , Katerina Chatzimeletiou7
, Katerina Chatzimeletiou7 

 , Grigoris Grimbizis7
, Grigoris Grimbizis7 

 , Basil Tarlatzis7
, Basil Tarlatzis7 

 , Maria Syrrou7
, Maria Syrrou7 

 , Alexandros Lambropoulos2
, Alexandros Lambropoulos2 




 , Socrates Katafigiotis2
, Socrates Katafigiotis2 

 , Theofanis Vavilis3
, Theofanis Vavilis3 

 , Zoe Papadopoulou4
, Zoe Papadopoulou4 

 , Giorgos Tzimagiorgis5
, Giorgos Tzimagiorgis5 

 , Anna-Bettina Haidich6
, Anna-Bettina Haidich6 

 , Katerina Chatzimeletiou7
, Katerina Chatzimeletiou7 

 , Grigoris Grimbizis7
, Grigoris Grimbizis7 

 , Basil Tarlatzis7
, Basil Tarlatzis7 

 , Maria Syrrou7
, Maria Syrrou7 

 , Alexandros Lambropoulos2
, Alexandros Lambropoulos2 


1- Laboratory of Genetics, 1st Department of Obstetrics and Gynecology, School of Medicine, Aristotle University of Thessaloniki, “Papageorgiou” General Hospital, Thessaloniki, Greece. , argyrakimar@gmail.com
2- Laboratory of Genetics, 1st Department of Obstetrics and Gynecology, School of Medicine, Aristotle University of Thessaloniki, “Papageorgiou” General Hospital, Thessaloniki, Greece.
3- Laboratory of Biology and Genetics, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
4- Laboratory of Biology, Faculty of Medicine, School of Health Sciences, University of Ioannina, Ioannina, Greece.
5- Laboratory of Biological Chemistry, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
6- Department of Hygiene and Epidemiology, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
7- Unit for Human Reproduction, 1st Department of Obstetrics and Gynecology, School of Medicine, Aristotle University of Thessaloniki, “Papageorgiou” General Hospital, Thessaloniki, Greece.
2- Laboratory of Genetics, 1st Department of Obstetrics and Gynecology, School of Medicine, Aristotle University of Thessaloniki, “Papageorgiou” General Hospital, Thessaloniki, Greece.
3- Laboratory of Biology and Genetics, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
4- Laboratory of Biology, Faculty of Medicine, School of Health Sciences, University of Ioannina, Ioannina, Greece.
5- Laboratory of Biological Chemistry, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
6- Department of Hygiene and Epidemiology, School of Medicine, Aristotle University of Thessaloniki, Thessaloniki, Greece.
7- Unit for Human Reproduction, 1st Department of Obstetrics and Gynecology, School of Medicine, Aristotle University of Thessaloniki, “Papageorgiou” General Hospital, Thessaloniki, Greece.
Full-Text [PDF 1213 kb]
(1054 Downloads)
| Abstract (HTML) (2021 Views)
Full-Text: (471 Views)
- Introduction
Vaginal delivery (VD) exposes infants to various stressors absent during caesarean section (CS), such as the secretion of catecholamines and cortisol, which trigger hormonal cascades in order to prepare the infant for life after birth (1). Furthermore, the artificial interventions employed during assisted reproductive technologies (ARTs), coincide with the embryos’ epigenetic reprogramming, thus leading possibly to aberrant establishment and maintenance of genomic imprints and an increase in imprinting disorders (2-5). On the other hand, concerning cellular stress, the octamer-binding transcription factor 4-B1 isoform (Oct4B1), has emerged as a possible cell stress marker (6). This isoform has been found to be an emergent marker of stemness in embryonic cells and is also implicated in apoptosis and probably in cancer (7-10). In the present study, Oct4B1 was selected to be studied because of its putative role in cellular stress response. This is the first study conducted in umbilical cord blood (UCB) concerning Oct4B1. Other genes susceptible to epigenetic modification upon stress exposure are Insulin Growth Factor 2 (IGF2), Mesoderm-Specific Transcript (MEST), and Paternally Expressed Gene 10 (PEG10). These genes affect fetal growth and their disruption is linked to metabolic disorders, cognitive impairment, low birth weight, and some types of cancer (11-13). These genes were selected to be studied because of their crucial role in fetal development, as they control embryonic and placental growth. Furthermore, these imprinted genes are responsive to different in utero environments and thus may mediate adverse environmental signals to the embryo during pregnancy. There are contradictory results in literature concerning the effect of ART on these genes. Lastly, there are no previous reports of the expression of these genes in embryos with breech presentation.
Given that various environmental factors and stressors can affect the epigenetic processes, concerns are raised about the implications of the increasing prevalence in elective CS deliveries and the use of ART. Our aim was to investigate the role of different modes of conception and delivery as possible environmental stressors on epigenetic regulation and stress response. Therefore, we evaluated the expression level of Oct4B1 and the expression and methylation status of IGF2, MEST, and PEG10 imprinted genes in UCB samples of newborns.
Given that various environmental factors and stressors can affect the epigenetic processes, concerns are raised about the implications of the increasing prevalence in elective CS deliveries and the use of ART. Our aim was to investigate the role of different modes of conception and delivery as possible environmental stressors on epigenetic regulation and stress response. Therefore, we evaluated the expression level of Oct4B1 and the expression and methylation status of IGF2, MEST, and PEG10 imprinted genes in UCB samples of newborns.
- Materials and Methods
- 1. Sampling method
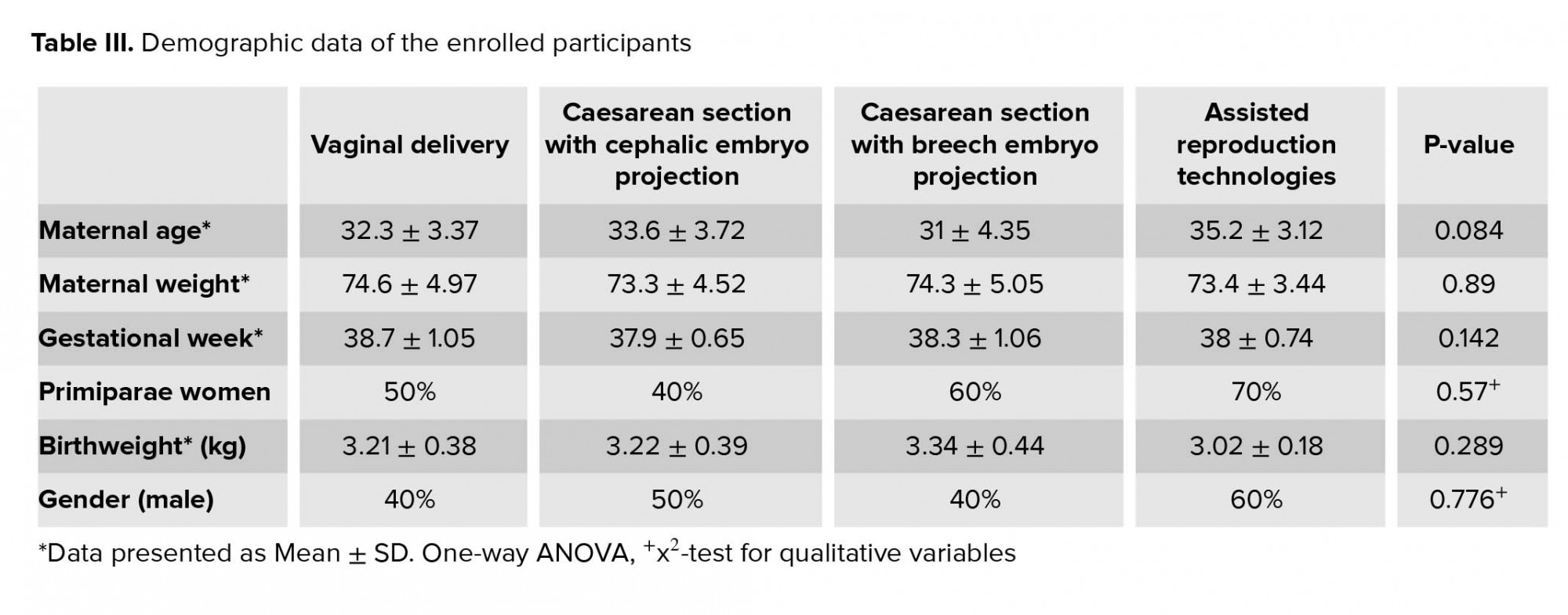
In this cohort study, 40 participants were enrolled whose UCB samples were collected during the time of their delivery. The samples were subdivided into four groups that comprised of 10 participants each. The extremely strict inclusion criteria did not allow for a larger sample size to be collected. This may be a disadvantage for the statistical analysis of the results, but it reflects a very homogenous and carefully designed sample population. The groups examined were VD (control group), CS with cephalic projection of the embryo, and CS with breech embryo projection. Additionally, the fourth group consisted of infants conceived through ART and delivered through caesarian section.
The eligibility for participation included uncomplicated full-term deliveries (≥ 37 wk) of healthy parents with an existing full medical record and supervision of the pregnancy by an obstetrician. On the other hand, the exclusion criteria included positive smoking status, chronic or acute diseases, twin gestations, non-compliance to the required medical tests during pregnancy, and pregnancies with embryos presenting chromosomal or anatomical abnormalities. All biospecimens collected were done so with the participant’s informed written consent after having secured permission from the Scientific Committee of Papageorgiou General Hospital of Thessaloniki, the Bioethics Committee of the School of Medicine of Aristotle University of Thessaloniki, and from the Hellenic Data Protection Authority.
The eligibility for participation included uncomplicated full-term deliveries (≥ 37 wk) of healthy parents with an existing full medical record and supervision of the pregnancy by an obstetrician. On the other hand, the exclusion criteria included positive smoking status, chronic or acute diseases, twin gestations, non-compliance to the required medical tests during pregnancy, and pregnancies with embryos presenting chromosomal or anatomical abnormalities. All biospecimens collected were done so with the participant’s informed written consent after having secured permission from the Scientific Committee of Papageorgiou General Hospital of Thessaloniki, the Bioethics Committee of the School of Medicine of Aristotle University of Thessaloniki, and from the Hellenic Data Protection Authority.
- 2. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from leucocytes using TRIZOL reagent (Invitrogen, USA), according to the manufacturer’s instructions. RNA samples were treated with TURBO DNA-free™ DNase (Ambion, Life Technologies, USA). A no-reverse transcription (no-RT) control was used for each sample to exclude any potential non-specific amplification of genomic DNA. The SuperScript™ First-Strand Synthesis System (Invitrogen, USA) was used to reverse transcribe total RNA with random hexamer primers. Quantitative Real-Time PCR employed TaqMan™ Gene Expression MasterMix reagent (Thermo Scientific, USA) and TaqMan™ Gene Expression Assays for evaluation of Beta-2 Microglobulin (B2M, endogenous control) IGF2, MEST, and PEG10 transcription. The primers for Oct4B1 were previously described (Table I). Quantification of the results was performed using the 2-ΔΔCt method after normalization against B2M.
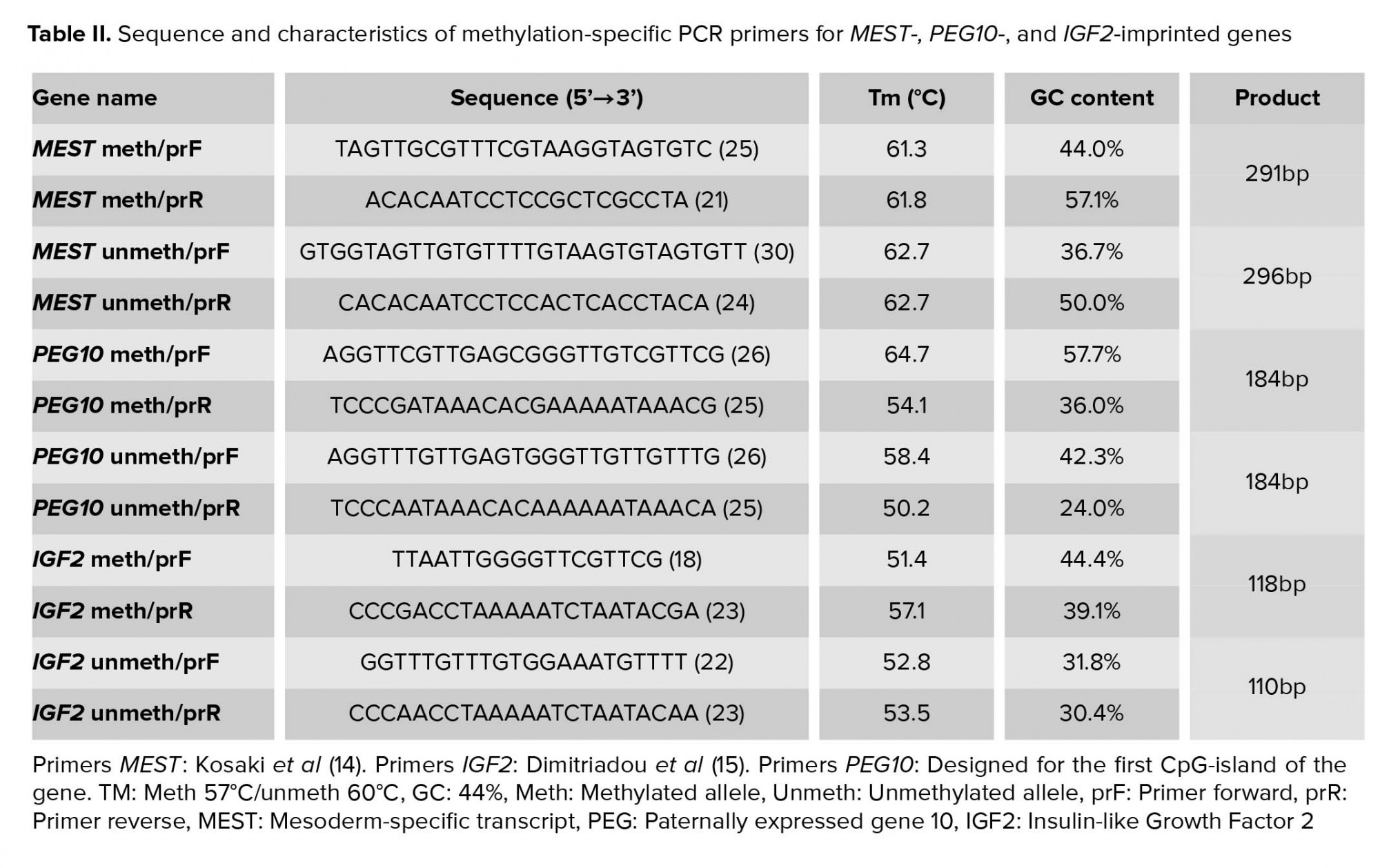
2.3. Methylation-specific PCR
For DNA extraction, DNA Mini Kit (Qiagen, Netherlands) was used. DNA recovered from the aforementioned process was chemically converted using EpiTectPlus DNA Bisulfite Kit (Qiagen, Netherlands), in order to turn unmethylated cytosines to uracil for detection by methylation-specific PCR. Next, the bisulfite-treated DNA was subjected to PCR for detection of the methylated and unmethylated MEST, IGF2, and PEG10 alleles (Table II). Samples containing no DNA were used as a negative control, whereas for positive control, bisulfite-treated DNA from peripheral blood was used.

2.4. Ethical considerations
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Scientific Committee of Papa Georgiou General Hospital of Thessaloniki (reference number: 157/27.06.12), the Bioethics Committee of the School of Medicine of Aristotle University of Thessaloniki(reference number: 1/8-11-2012), and of the Hellenic Data Protection Authority (reference number: 1145) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. In addition, a written informed consent was obtained from individual participants included in the study.
2.5. Statistical analysis
Data were analyzed using the IBM SPSS Statistics version.24 (SPSS Inc., Chicago, Illinois, USA) program. Demographic data were normally distributed (based on Shapiro test for normality) and one-way ANOVA-test was used for quantitative variables (maternal age, maternal weight, gestational week, and birth weight) and χ2-test for qualitative variables (gender of the newborns and ratio of primiparae women). The gene expression results were evaluated through non-parametrical tests due to the small sample size, and more specifically the Kruskal-Wallis test was employed to compare Oct4B1, IGF2, MEST, and PEG10 expression between the four groups. Results were deemed significant only when p < 0.05.
3.Results
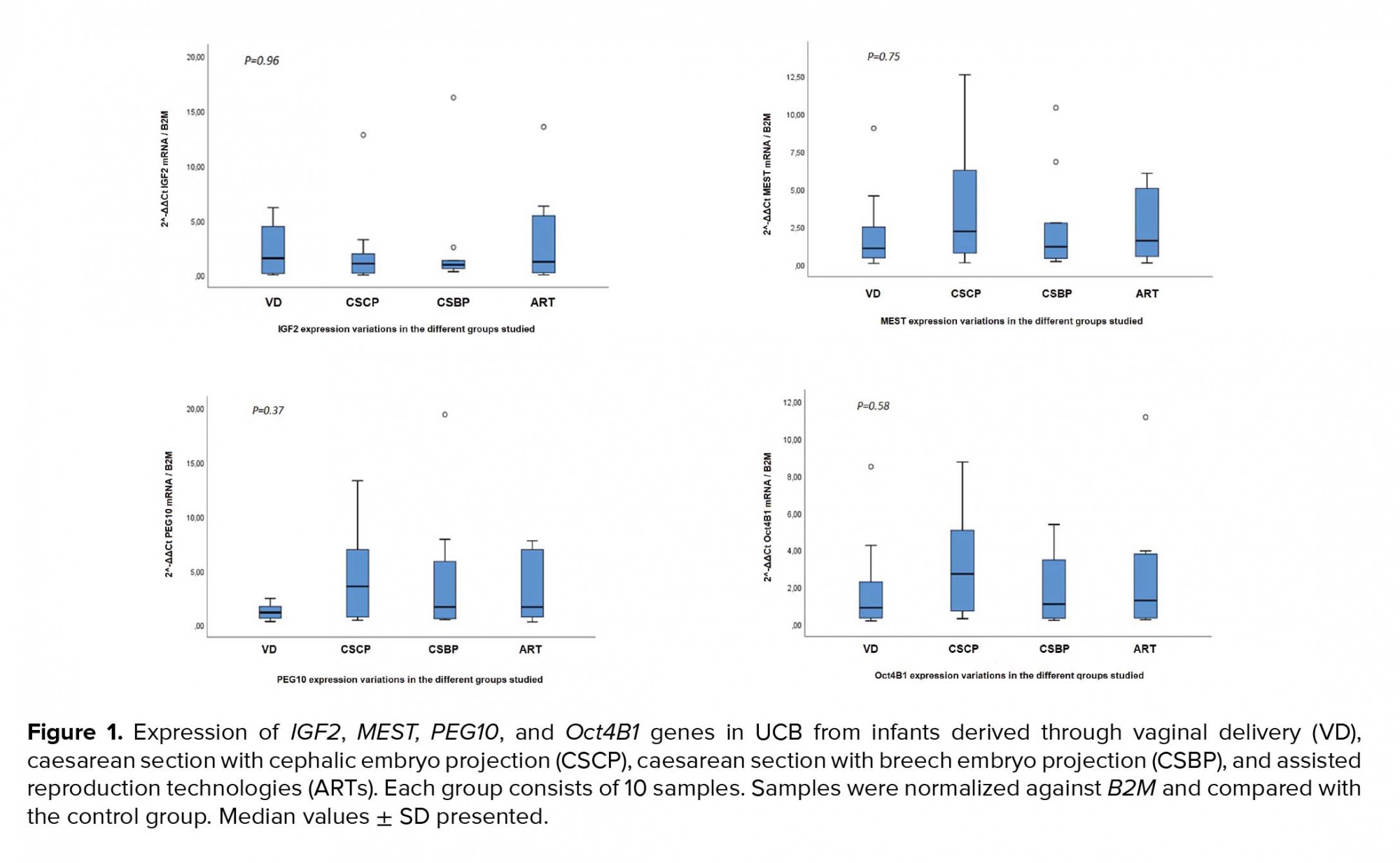
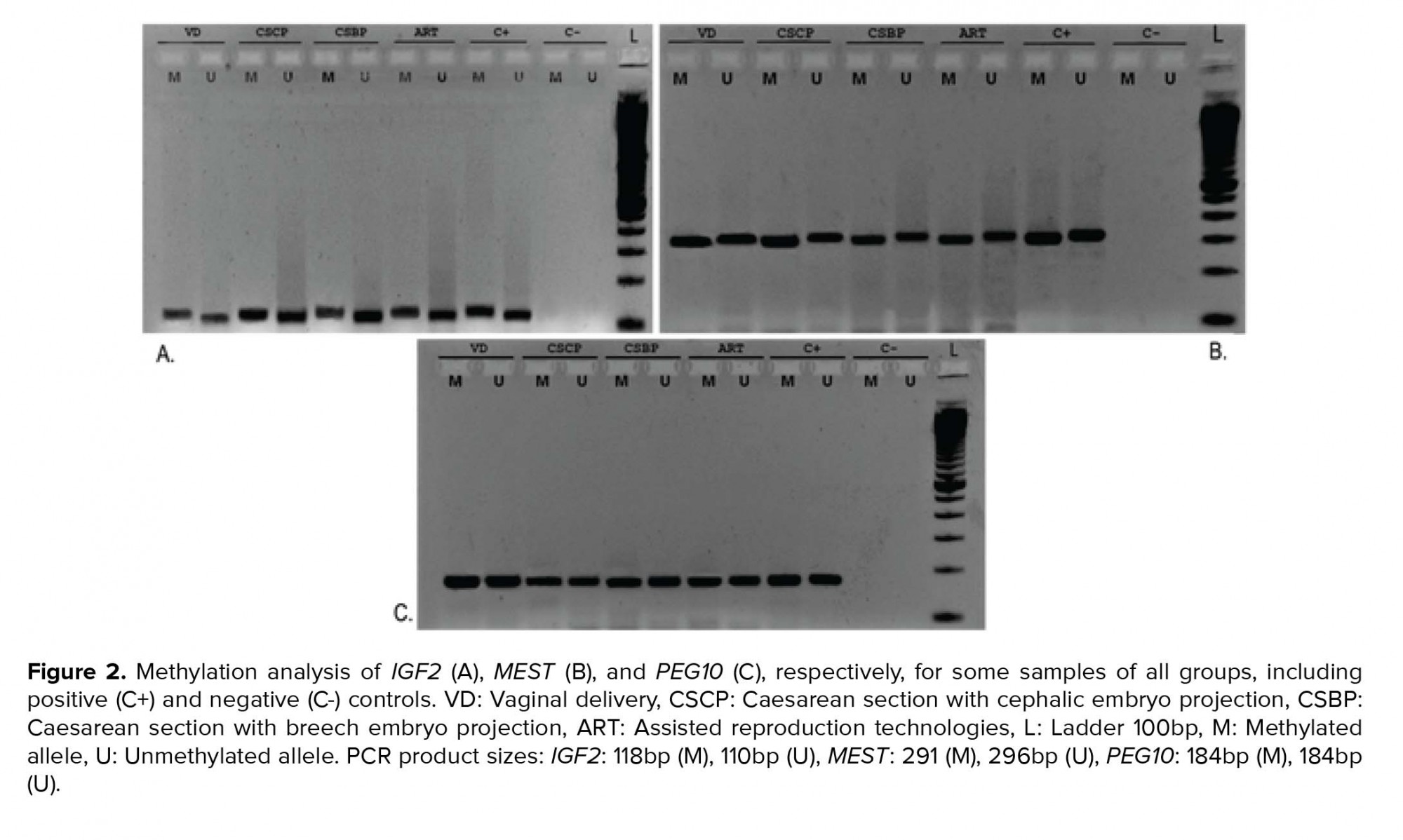
As mentioned above, 40 participants were enrolled whose UCB samples were collected during the time of their delivery (Table III). Quantitative Real-time PCR revealed no statistically significant differences at the transcriptional level between the VD, CS, and ART groups for Oct4B1 (p = 0.58), IGF2 (p = 0.96), MEST (p = 0.75), and PEG10 (p = 0.37) (Figure 1). PEG10 exhibited a non-significant trend for up regulation (p = 0.089) in the group of cephalic presentation CS compared to the VD group. Methylation-Specific PCR products were subjected to gel electrophoresis in order to validate the presence of both methylated and unmethylated alleles for IGF2, MEST, and PEG10 for all 40 samples (Figure 2). Methylation analysis was not possible for Oct4B1 due to the existence of alternative promoters for this isoform, incommoding primer design (16). Methylation status is in agreement with non-statistical differences in expression.
4. Discussion
It is assumed that imprinted genes can serve as environmental sensors, since they can be responsive to environmental stressors and alter their expression and/or methylation during specific developmental stages. In the current study, we investigated the effect of the conception and delivery mode, which are considered potential environmental stressors, on the expression and methylation levels of three imprinted genes. Furthermore, we evaluated their effect on the expression of Oct4B1isoform, a potential stress-response marker. In our study, it appears that different modes of conception and delivery do not alter significantly the mRNA expression levels for Oct4B1, IGF2, MEST, and PEG10 in UCB. This could be attributed not only to the small sample size available, but also to the extremely strict inclusion criteria, excluding any kind of fetal abnormality, complicated pregnancies, mothers with diagnosed health conditions, and unsupervised pregnancies.
To date, to the best of our knowledge, no other reports exist in literature investigating the expression levels of the Oct4B1 isoform in UCB. Oct4B1 is not only a newly discovered variant of Oct4 family that bestows stemness qualities in cell expressing it, but also seems to serve an anti-apoptotic function in a multitude of cancer cell lines (9, 10, 17, 18). Furthermore, Oct4B1 warrants further investigation as a regulator of cellular stress response since it plays a role in the regulation of HSP40 family of chaperones (17). The implications of this regulation are not clear at the moment, but it is possible that Oct4B1 might modulate globally the stress response since the HSP40 family constitutes the most diverse and largest category of HSPs and its members are known to interact with other actively researched HSP families such as HSP70 and HSP90 (19). Different stress conditions during CS and ART could probably result in alterations in Oct4B1 expression levels. However, this was not observed in our study, indicating that the stress variations during different conception and delivery modes are not capable of affectingOct4B1 expression.
Furthermore, this is the first study on the expression and methylation of those genes that correlate them to embryo presentation during CS, either cephalic or breech. Such a distinction is of importance since the positioning of the embryo can indicate maturity prior to delivery and either excessive or suboptimal levels of stress could possibly interfere with methylation and future health outcomes. Interestingly, in our study, there was a trend for increased expression of PEG10 in the CSCP group, a finding that warrants further investigation in broader sample studies. As far as the ART group is concerned, our results are consistent with previous studies, indicating stability in the expression and DNA methylation levels of the aforementioned imprinted genes. However, there are other studies reporting altered DNA methylation and expression levels of these genes after ART treatments. These contradictory results can be attributed to different ART protocols, variation in sample size, sample homogeneity, and different analysis methods. Furthermore, there are also other mechanisms that maintain imprints, such as histone modifications and long non-coding RNAs, which should also be considered as a possible explanation to the variation of results found between studies. It remains also unclear whether the parental underlying sub-fertility per se can act as the causal factor of the higher complication prevalence observed in the results of some reports of ART-conceived children. Lastly, the existence of an imprinted gene network could also explain the heterogeneity of the reported results. This theory was first described in animal reports (20, 21), but many scientists believe that it also refers to humans (22-24). According to this theory, the imprinted gene network includes hundreds of imprinted genes with similar roles controlling essential functions, such as fetal nutrition and growth. Thus, the inappropriate function of some imprinted genes can be compensated by the expression of others with similar role.
All evidence shows that it is necessary to understand the health risks and the underlying molecular mechanisms of ART interventions in order to increase the safety of these techniques. Thus, the contradictory literature results call for caution and for larger studies. The current study is exploratory in nature, utilizing well-characterized groups as far as the medical history of the participants and the medical supervision during pregnancy is concerned. It can be considered though as a starting point for more resolving the role of imprinted genes and Oct4B1 isoform on the regulation of human reproduction. Due to the limitation of the sample size, further studies are needed to evaluate these results.
5. Conclusion
The results indicate that different modes of conception and delivery do not affect the mRNA expression levels of Oct4B1, IGF2, MEST, and PEG10 in UCB. These results are the first reported in cephalic and breech presentation CS concerning these specific genes. Additionally, methylation analysis revealed the presence of both methylated and unmethylated alleles for the above aforementioned imprinted genes, with no methylation differences reported between groups. Methylation status of all genes is in consistence with non-statistical differences in expression. Methylation analysis was not possible for Oct4B1 due to the existence of unknown alternative promoters for this isoform. While our study found no effect of the mode of conception and delivery on the expression of Oct4B1, IGF2, MEST, and PEG10 in UCB, the expression of the genes in question should be investigated further. As far as the ART group is concerned, our results are consistent with some previous reports. However, there are other studies reporting altered DNA methylation and expression of these genes after ART manipulations. These contradictory literature results call for caution and for larger studies. Differences may become apparent utilizing larger sample sizes, keeping though in mind that excellent characterization of the groups examined (medical history, supervision, etc.) should not be compromised for achieving the desired sample size. This would ensure that the observed effect can be strongly correlated to the mode of conception and delivery, rather than confounding variables.
Acknowledgements
The authors thank “Papageorgiou” Foundation and the 1st Department of Obstetrics and Gynecology for funding the research. They are also thankful to the participants who took part in this study, as well as all medical staff of Papageorgiou’s Hospital 1st Obstetrics and Gynecology University Clinic who helped in the collection of the samples.
Conflict of interest
During the recent five-yr, Basil C. Tarlatzis has received honoraria, fees, or grant support from the following organizations(in alphabetic order): DIAFERT, European :union: Social Fund and the Greek Ministry of Education, European :union: and the Greek General Secretariat of Research and Technology, Ferring, IBSA, MERCK, MSD, Ova science, Roche Diagnostics. The other authors declare no conflict of interest.
2.3. Methylation-specific PCR
For DNA extraction, DNA Mini Kit (Qiagen, Netherlands) was used. DNA recovered from the aforementioned process was chemically converted using EpiTectPlus DNA Bisulfite Kit (Qiagen, Netherlands), in order to turn unmethylated cytosines to uracil for detection by methylation-specific PCR. Next, the bisulfite-treated DNA was subjected to PCR for detection of the methylated and unmethylated MEST, IGF2, and PEG10 alleles (Table II). Samples containing no DNA were used as a negative control, whereas for positive control, bisulfite-treated DNA from peripheral blood was used.


2.4. Ethical considerations
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Scientific Committee of Papa Georgiou General Hospital of Thessaloniki (reference number: 157/27.06.12), the Bioethics Committee of the School of Medicine of Aristotle University of Thessaloniki(reference number: 1/8-11-2012), and of the Hellenic Data Protection Authority (reference number: 1145) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. In addition, a written informed consent was obtained from individual participants included in the study.
2.5. Statistical analysis
Data were analyzed using the IBM SPSS Statistics version.24 (SPSS Inc., Chicago, Illinois, USA) program. Demographic data were normally distributed (based on Shapiro test for normality) and one-way ANOVA-test was used for quantitative variables (maternal age, maternal weight, gestational week, and birth weight) and χ2-test for qualitative variables (gender of the newborns and ratio of primiparae women). The gene expression results were evaluated through non-parametrical tests due to the small sample size, and more specifically the Kruskal-Wallis test was employed to compare Oct4B1, IGF2, MEST, and PEG10 expression between the four groups. Results were deemed significant only when p < 0.05.
3.Results
As mentioned above, 40 participants were enrolled whose UCB samples were collected during the time of their delivery (Table III). Quantitative Real-time PCR revealed no statistically significant differences at the transcriptional level between the VD, CS, and ART groups for Oct4B1 (p = 0.58), IGF2 (p = 0.96), MEST (p = 0.75), and PEG10 (p = 0.37) (Figure 1). PEG10 exhibited a non-significant trend for up regulation (p = 0.089) in the group of cephalic presentation CS compared to the VD group. Methylation-Specific PCR products were subjected to gel electrophoresis in order to validate the presence of both methylated and unmethylated alleles for IGF2, MEST, and PEG10 for all 40 samples (Figure 2). Methylation analysis was not possible for Oct4B1 due to the existence of alternative promoters for this isoform, incommoding primer design (16). Methylation status is in agreement with non-statistical differences in expression.



4. Discussion
It is assumed that imprinted genes can serve as environmental sensors, since they can be responsive to environmental stressors and alter their expression and/or methylation during specific developmental stages. In the current study, we investigated the effect of the conception and delivery mode, which are considered potential environmental stressors, on the expression and methylation levels of three imprinted genes. Furthermore, we evaluated their effect on the expression of Oct4B1isoform, a potential stress-response marker. In our study, it appears that different modes of conception and delivery do not alter significantly the mRNA expression levels for Oct4B1, IGF2, MEST, and PEG10 in UCB. This could be attributed not only to the small sample size available, but also to the extremely strict inclusion criteria, excluding any kind of fetal abnormality, complicated pregnancies, mothers with diagnosed health conditions, and unsupervised pregnancies.
To date, to the best of our knowledge, no other reports exist in literature investigating the expression levels of the Oct4B1 isoform in UCB. Oct4B1 is not only a newly discovered variant of Oct4 family that bestows stemness qualities in cell expressing it, but also seems to serve an anti-apoptotic function in a multitude of cancer cell lines (9, 10, 17, 18). Furthermore, Oct4B1 warrants further investigation as a regulator of cellular stress response since it plays a role in the regulation of HSP40 family of chaperones (17). The implications of this regulation are not clear at the moment, but it is possible that Oct4B1 might modulate globally the stress response since the HSP40 family constitutes the most diverse and largest category of HSPs and its members are known to interact with other actively researched HSP families such as HSP70 and HSP90 (19). Different stress conditions during CS and ART could probably result in alterations in Oct4B1 expression levels. However, this was not observed in our study, indicating that the stress variations during different conception and delivery modes are not capable of affectingOct4B1 expression.
Furthermore, this is the first study on the expression and methylation of those genes that correlate them to embryo presentation during CS, either cephalic or breech. Such a distinction is of importance since the positioning of the embryo can indicate maturity prior to delivery and either excessive or suboptimal levels of stress could possibly interfere with methylation and future health outcomes. Interestingly, in our study, there was a trend for increased expression of PEG10 in the CSCP group, a finding that warrants further investigation in broader sample studies. As far as the ART group is concerned, our results are consistent with previous studies, indicating stability in the expression and DNA methylation levels of the aforementioned imprinted genes. However, there are other studies reporting altered DNA methylation and expression levels of these genes after ART treatments. These contradictory results can be attributed to different ART protocols, variation in sample size, sample homogeneity, and different analysis methods. Furthermore, there are also other mechanisms that maintain imprints, such as histone modifications and long non-coding RNAs, which should also be considered as a possible explanation to the variation of results found between studies. It remains also unclear whether the parental underlying sub-fertility per se can act as the causal factor of the higher complication prevalence observed in the results of some reports of ART-conceived children. Lastly, the existence of an imprinted gene network could also explain the heterogeneity of the reported results. This theory was first described in animal reports (20, 21), but many scientists believe that it also refers to humans (22-24). According to this theory, the imprinted gene network includes hundreds of imprinted genes with similar roles controlling essential functions, such as fetal nutrition and growth. Thus, the inappropriate function of some imprinted genes can be compensated by the expression of others with similar role.
All evidence shows that it is necessary to understand the health risks and the underlying molecular mechanisms of ART interventions in order to increase the safety of these techniques. Thus, the contradictory literature results call for caution and for larger studies. The current study is exploratory in nature, utilizing well-characterized groups as far as the medical history of the participants and the medical supervision during pregnancy is concerned. It can be considered though as a starting point for more resolving the role of imprinted genes and Oct4B1 isoform on the regulation of human reproduction. Due to the limitation of the sample size, further studies are needed to evaluate these results.
5. Conclusion
The results indicate that different modes of conception and delivery do not affect the mRNA expression levels of Oct4B1, IGF2, MEST, and PEG10 in UCB. These results are the first reported in cephalic and breech presentation CS concerning these specific genes. Additionally, methylation analysis revealed the presence of both methylated and unmethylated alleles for the above aforementioned imprinted genes, with no methylation differences reported between groups. Methylation status of all genes is in consistence with non-statistical differences in expression. Methylation analysis was not possible for Oct4B1 due to the existence of unknown alternative promoters for this isoform. While our study found no effect of the mode of conception and delivery on the expression of Oct4B1, IGF2, MEST, and PEG10 in UCB, the expression of the genes in question should be investigated further. As far as the ART group is concerned, our results are consistent with some previous reports. However, there are other studies reporting altered DNA methylation and expression of these genes after ART manipulations. These contradictory literature results call for caution and for larger studies. Differences may become apparent utilizing larger sample sizes, keeping though in mind that excellent characterization of the groups examined (medical history, supervision, etc.) should not be compromised for achieving the desired sample size. This would ensure that the observed effect can be strongly correlated to the mode of conception and delivery, rather than confounding variables.
Acknowledgements
The authors thank “Papageorgiou” Foundation and the 1st Department of Obstetrics and Gynecology for funding the research. They are also thankful to the participants who took part in this study, as well as all medical staff of Papageorgiou’s Hospital 1st Obstetrics and Gynecology University Clinic who helped in the collection of the samples.
Conflict of interest
During the recent five-yr, Basil C. Tarlatzis has received honoraria, fees, or grant support from the following organizations(in alphabetic order): DIAFERT, European :union: Social Fund and the Greek Ministry of Education, European :union: and the Greek General Secretariat of Research and Technology, Ferring, IBSA, MERCK, MSD, Ova science, Roche Diagnostics. The other authors declare no conflict of interest.
Type of Study: Short Research Reports |
Subject:
Reproductive Biology
References
1. Lagercrantz H. The good stress of being born. Acta Paediatr 2016; 105: 1413-1416. [DOI:10.1111/apa.13615] [PMID]
2. Anifandis G, Messini CI, Dafopoulos K, Messinis IE. Genes and conditions controlling mammalian pre- and post-implantation embryo development. Curr Genomics 2015; 16: 32-46. [DOI:10.2174/1389202916666141224205025] [PMID] [PMCID]
3. Canovas S, Ross PJ, Kelsey G, Coy P. DNA methylation in embryo development: Epigenetic impact of ART (assisted reproductive technologies). Bioessays 2017; 39: 1700106: 1-11. [DOI:10.1002/bies.201700106] [PMID]
4. Chen J, Li Q, Rialdi A, Mystal E, Ly J, Finik J, et al. Influences of maternal stress during pregnancy on the epi/genome: comparison of placenta and umbilical cord blood. J Depress Anxiety 2014; 3: 152-167.
5. Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T, et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta 2015; 36: 1100-1105. [DOI:10.1016/j.placenta.2015.08.008] [PMID]
6. Takeda J, Seino S, Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res 1992; 20: 4613-4620. [DOI:10.1093/nar/20.17.4613] [PMID] [PMCID]
7. Farashahi Yazd E, Rafiee MR, Soleimani M, Tavallaei M, Kabir Salmani M, Mowla SJ. OCT4B1, a novel spliced variant of OCT4, generates a stable truncated protein with a potential role in stress response. Cancer Lett 2011; 309:170-175. [DOI:10.1016/j.canlet.2011.05.027] [PMID]
8. Gao Y, Wei J, Han J, Wang X, Su G, Zhao Y, et al. The novel function of OCT4B isoform-265 in genotoxic stress. Stem Cells 2012; 30: 665-672. [DOI:10.1002/stem.1034] [PMID]
9. Papamichos SI, Kotoula V, Tarlatzis BC, Agorastos T, Papazisis K, Lambropoulos AF.OCT4B1 isoform: the novel OCT4 alternative spliced variant as a putative marker of stemness. Mol Hum Reprod 2009; 15: 269-270. [DOI:10.1093/molehr/gap018] [PMID]
10. Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000; 24: 372-376. [DOI:10.1038/74199] [PMID]
11. St-Pierre J, Hivert MF, Perron P, Poirier P, Guay SP, Brisson D, et al. IGF2 DNA methylation is a modulator of newborn's fetal growth and development. Epigenetics 2012; 7: 1125-1132. [DOI:10.4161/epi.21855] [PMID] [PMCID]
12. Kappil MA, Green BB, Armstrong DA, Sharp AJ, Lambertini L, Marsit CJ, et al. Placental expression profile of imprinted genes impacts birth weight. Epigenetics 2015; 10: 842-849. [DOI:10.1080/15592294.2015.1073881] [PMID] [PMCID]
13. Lim AL, Ng S, Leow SC, Choo R, Ito M, Chan YH, et al. Epigenetic state and expression of imprinted genes in umbilical cord correlates with growth parameters in human pregnancy. J Med Genet 2012; 49:689-697. [DOI:10.1136/jmedgenet-2012-100858] [PMID]
14. Kosaki K, Kosaki R, Craigen WJ, Matsuo N. Isoform-specific imprinting of the human PEG1/MEST gene. Am J Hum Genet 2000; 66: 309-312. [DOI:10.1086/302712] [PMID] [PMCID]
15. Dimitriadou E, Noutsopoulos D, Markopoulos G, Vlaikou AM, Mantziou S, Traeger-Synodinos J, et al. Abnormal DLK1/MEG3 imprinting correlates with decreased HERV-K methylation after assisted reproduction and preimplantation genetic diagnosis. Stress 2013; 16: 689-697. [DOI:10.3109/10253890.2013.817554] [PMID]
16. Papamichos SI, Lambropoulos AF, Kotoula V. OCT4B expression in PBMNCs suggests the existence of an alternative OCT4 promoter. Genes Chromosomes Cancer 2009; 48: 1112-1114. [DOI:10.1002/gcc.20707] [PMID]
17. Mirzaei MR, Asadi MH, Mowla SJ, Hassanshahi G, Ahmadi Z. Down-regulation of HSP40 gene family following OCT4B1 suppression in human tumor cell lines. Iran J Basic Med Sci 2016; 19: 187-193.
18. Asadzadeh J, Asadi MH, Shakhssalim N, Rafiee MR, Kalhor HR, Tavallaei M, et al. A plausible anti-apoptotic role of up-regulated OCT4B1 in bladder tumors. Urol J 2012; 9: 574-580.
19. Poursani EM, Mehravar M, Shahryari A, Mowla SJ, Soltani BM. Alternative splicing generates different 5' UTRs in OCT4B Variants. Avicenna J Med Biotechnol 2017; 9: 201-204.
20. Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 2006; 11: 711-722. [DOI:10.1016/j.devcel.2006.09.003] [PMID]
21. Fauque P, Ripoche MA, Tost J, Journot L, Gabory A, Busato F, et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet 2010; 19: 1779-1790. [DOI:10.1093/hmg/ddq059] [PMID]
22. Choux C, Carmignac V, Bruno C, Sagot P, Vaiman D, Fauque P. The placenta: Phenotypic and epigenetic modifications induced by assisted reproductive technologies throughout pregnancy. Clin Epigenetics 2015; 7: 87-106. [DOI:10.1186/s13148-015-0120-2] [PMID] [PMCID]
23. Choux C, Binquet C, Carmignac V, Bruno C, Chapusot C, Barberet J,et al. The epigenetic control of transposable elements and imprinted genes in newborns is affected by the mode of conception: ART versus spontaneous conception without underlying infertility. Hum Reprod 2018; 33: 331-340. [DOI:10.1093/humrep/dex366] [PMID]
24. Iglesias-Platas I, Martin-Trujillo A, Petazzi P, Guillaumet-Adkins A, Esteller M, Monk D. Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum Mol Genet 2014; 23: 6275-6285. [DOI:10.1093/hmg/ddu347] [PMID] [PMCID]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





