Wed, Apr 2, 2025
[Archive]
Volume 19, Issue 1 (January 2021)
IJRM 2021, 19(1): 23-34 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Najafipour R, Momeni A, Yousefipour F, Mousavi S, Moghbelinejad S. Underexpression of hsa-miR-449 family and their promoter hypermethylation in infertile men: A case-control study. IJRM 2021; 19 (1) :23-34
URL: http://ijrm.ir/article-1-1628-en.html
URL: http://ijrm.ir/article-1-1628-en.html
Reza Najafipour1 

 , Abdolmabood Momeni2
, Abdolmabood Momeni2 

 , Farideh Yousefipour3
, Farideh Yousefipour3 

 , Shaghayegh Mousavi4
, Shaghayegh Mousavi4 

 , Sahar Moghbelinejad *
, Sahar Moghbelinejad * 

 5
5


 , Abdolmabood Momeni2
, Abdolmabood Momeni2 

 , Farideh Yousefipour3
, Farideh Yousefipour3 

 , Shaghayegh Mousavi4
, Shaghayegh Mousavi4 

 , Sahar Moghbelinejad *
, Sahar Moghbelinejad * 

 5
5
1- Research Institute for Prevention of Non-Communicable Diseases, Cellular and Molecular Research Centre, Qazvin University of Medical Sciences, Qazvin, Iran.
2- Biology Department, School of Basic Science, Arak University, Arak, Iran.
3- National Institute of Engineering and Biotechnology, Tehran, Iran.
4- Department of Molecular Medicine, School of Medicine, Qazvin University of Medical Sciences, Qazvin, Iran.
5- Research Institute for Prevention of Non-Communicable Diseases, Cellular and Molecular Research Centre, Qazvin University of Medical Sciences, Qazvin, Iran. ,smoghbelinejad@qums.ac.ir
2- Biology Department, School of Basic Science, Arak University, Arak, Iran.
3- National Institute of Engineering and Biotechnology, Tehran, Iran.
4- Department of Molecular Medicine, School of Medicine, Qazvin University of Medical Sciences, Qazvin, Iran.
5- Research Institute for Prevention of Non-Communicable Diseases, Cellular and Molecular Research Centre, Qazvin University of Medical Sciences, Qazvin, Iran. ,
Full-Text [PDF 16835 kb]
(943 Downloads)
| Abstract (HTML) (1928 Views)
1. Introduction
On average, it is estimated that infertility occurs in 10 to 15% of couples and 50% of infertility cases are due to male factor. The causes of 65-70% of male infertility cases is unknown and the correct mechanism has not been defined (1, 2). Genetic factors can be one of the causes of male infertility (3). But the role of miRNAs in the process of spermatogenesis and male infertility is very important. MiRNAs are non-coding RNAs, and play an important role in regulating gene expression (4, 5). MiRNAs regulate gene expression in two ways: by suppressing transcription and translation (RNAi) (6) or by activating transcription (RNAa) (7-9). The expression of miRNAs was shown in some types of male germ cells (10-14). The hsa-miR-449 was first detected in the fetal brain of mice(15, 16).
The hsa-miR-449 family has three members in mice and human, namely hsa-miR-449a, hsa-miR-449b, and hsa-miR-449c. These miRNAs have conserved sequence among different species and are located on the second intron of the Cdc20b gene. While the three hsa-miR-499 (miR-499 a, b, c) members have the same seed sequences, hsa-miR-449 members play the main role in the control of cell cycle and differentiation of epidermis (17-19).
In this regard, studies have shown that the hsa-miR449 family (hsa-miR-449a, hsa-miR449b, and hsa-miR-449c) and the hsa-miR-34 b, c (hsa-miR-34b-3p and hsa-miR-34c-5p) contain an identical seed sequence and have same sequence with the another miRNA, hsa-miR-34a-5p (17-21). About in addition, with respect to the role of hsa-miR-449 in spermatogenesis and male infertility, it was shown that the hsa-miR-449 family highly express in spermatocyte, spermatid, and adult testis. In one study was reported that, inactivation of both the hsa-miR34-b,c and hsa-miR-449 causes low sperm counts, motility and high abnormal sperm morphology in animal models (22). CpG methylation of mentioned genes is one possible reason for the their underexpression (20, 22), so that, under expression and high methylation of hsa-miR-449 (a, b, c) (as an important tumor-suppressor gene) have been shown in various cancers (23, 24). Besides, methylation of their promoter region has also been shown to be one of the mechanisms of expression reduction in adition to playing an important role in carcinogenesis of these microRNAs. Up to now, the patterns of hsa-miR-449 family expression and methylation has not been reported in different groups of infertile men and all the information in this regard has been based on animal models (25). Therefore, in this study we evaluated the expression and methylation pattern of hsa-miR-449 family in infertile men.
2. Materials and Methods
2.1. Subject recruitment and sampling
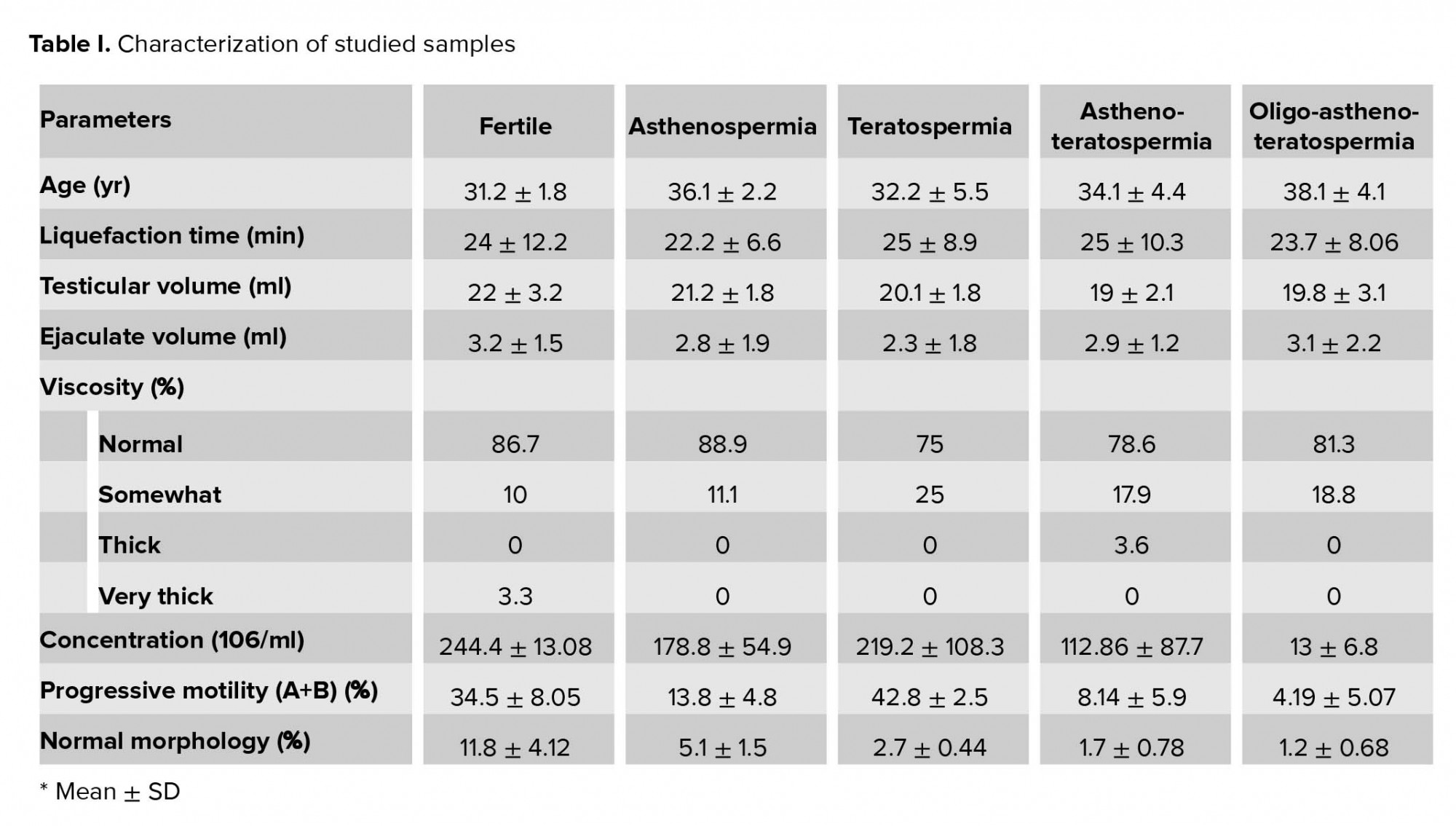
In this case-control study, 74 infertile men with idiopathic asthenozoospermia (n = 14), teratozoospermia (n = 16), asthenoteratozoospermia (n = 28), and oligoasthenoteratozoospermia (n = 16) were collected during 2018-2019 from the ACECR Telemedicine Infertility Center Qazvin, IRAN, based on the WHO criteria. The condition of infertile men was as follows: a history of infertility for at least 1 yr with their wives having a normal gynecological evaluation. However, infertile men conditions such as cystic fibrosis, Klinefelter syndrome, varicocele, chemotherapy, AZF, and genes micro deletions were not included in this study. In addition, 30 fertile healthy men were recruited as the control group. A questionnaire was designed to evaluation of the patients and controls’ information, including medical history, occupational and environmental condition, smoking condition (an adult who has smoked 100 cigarettes in his lifetime and who currently smokes cigarettes)and reproduction status (Table I). Patients were advised not to have sexual abstinence for three days before sampling. After sampling, semen samples were stored at 37ºC for 30 min to complete the liquefaction. Then, based on WHO criteria sperms concentration, motility, and morphology were evaluated (26).
2.2. RNA extraction and qRT-PCR
After liqufication, we centrifuged the semen samples for 10 min at 500 g. Then 1 ml FSB (Merck, Germany) was added to sperms pellet and somatic cells removed.TRIZOL reagent was used for isolation of the total RNA from the sperms based on kit protocols (Invitrogen Life Technology Co., USA). hsa-miR-449-a (MI0001648), hsa-miR-449-b (MI0003673), and hsa-449-c (MI0003823) were studied in this research. hsa-miR30a-5p (MIMAT0000087) and hsa-miR100-5p (MIMAT 0000102) were used as internal controls. Rotor gene-Q real-time PCR system (Qiagene, Germany) was used to quantifing of the RNA expression. Total amount of master mix was 10 µl and included 1 µl of reverse and forward primers (Exiqon, Denmark), 5 µl of Ampliqon real Q plus 2× master mix green (Ampliqone, Denmark), and 4 µl of diluted cDNA. For enzyme activation we incubated master mix for 15 min at 95ºC. Then reaction was runned in 40 cycle for 20 sec at 95ºC and 60 sec at 60ºC. Ct values was used for evaluation of expression rate of studied miRNAs. hsa-miR30a-5p and hsa-miR100-5p were used as the endogenous controls. The 2-△Ct method used for expression rate detection of target genes in comparision to internal controls.
2.3. DNA extraction and bisulfite modification
Phenol-chloroform method was used for DNA extraction. 2-5 µg of extracted DNA was bisulfited by using of EpiJET™ Bisulfite Conversion kit (Thermo Fisher Scientific, Inc).
2.4. MSPCR
The hsa-miR-449 methylation status was evaluated in all studied samples. For the targeted site, methylation-specific primers were designed. While the methylated primers were Forward: 5’-CGTTCGTTAATTTTTTCGTTTTTTGTCGC-3’) and Reverse: 5ꞌ-GTCAAAACCCGAATAAAATTCCCCGACG-3’, the unmethylated primers were Forward: 5’-TTGTTTGTTAATTTTTTTGTTTTTTGTTGT-3’ and Reverse: 5’- ATCAAAACCCAAATAAAATTCCCCAACA-3’. Methylation-specific PCR (MSP) was used to evaluation of methylation status of hsa-miR-449-abc promoter region. 1 µL of bisulfit converted DNA with methylated and unmethylated primers was amplifiedin in final 10 µL reaction mixture. "The PCR conditions for methylation status were: 95ºC for 15 min (Hot start), followed by 35 cycles at 95ºC for 20 sec (denaturation), 56.5ºC for 45 sec (annealing), and 72ºC for 45 sec (extension). PCR condition for unmethylated- primers was the same as the methylated condition except for the number of cycles and annealing temperature (60ºC)" (27).
2.5. Ethical considerations
This case-control study was approved by the Ethics Committee of Qazvin university of medical science with dedicated ID IR.QUMS.REC.1396.294. The recruited patients gave their informed written consent.
2.6. Statistical analysis
The GraphPad software (GraphPad PRISM V 5.04) was used for the data analysis. The analysis of variance test (ANOVA) was used for the evaluation of the miRNAs expression levels difference among the different studied groups. The frequency of promoter methylation pattern was evaluated by a nonparametric test (Kruskal-Wallis). The correlation between the miRNA expression rate, methylation with different sperm parameters was analyzed by Spearman's rank correlation. All P-values were two-tailed, with p < 0.05 considered as statistically significant.
3. Results
3.1. Expression of miRNAs in sperm samples of studied groups
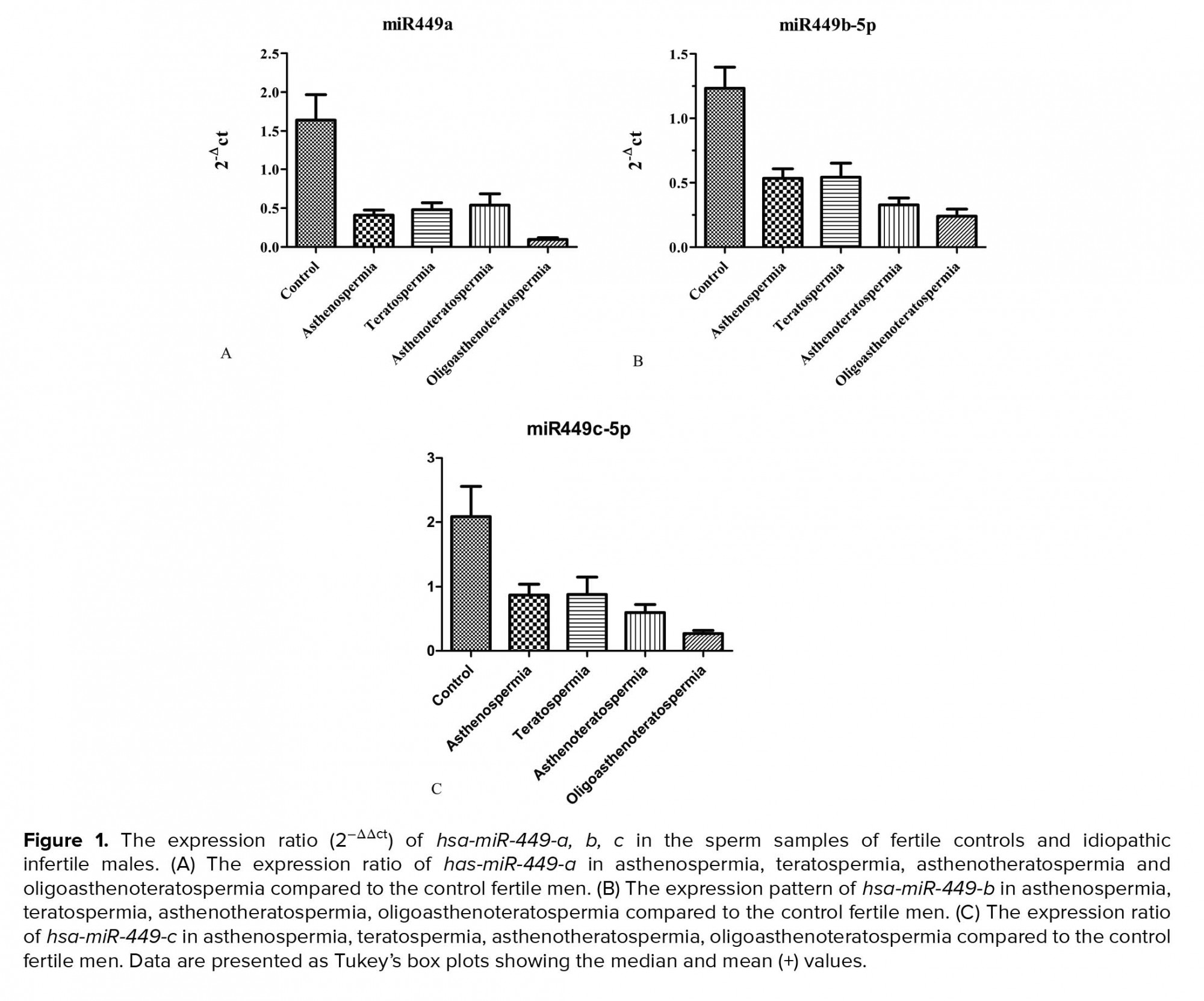
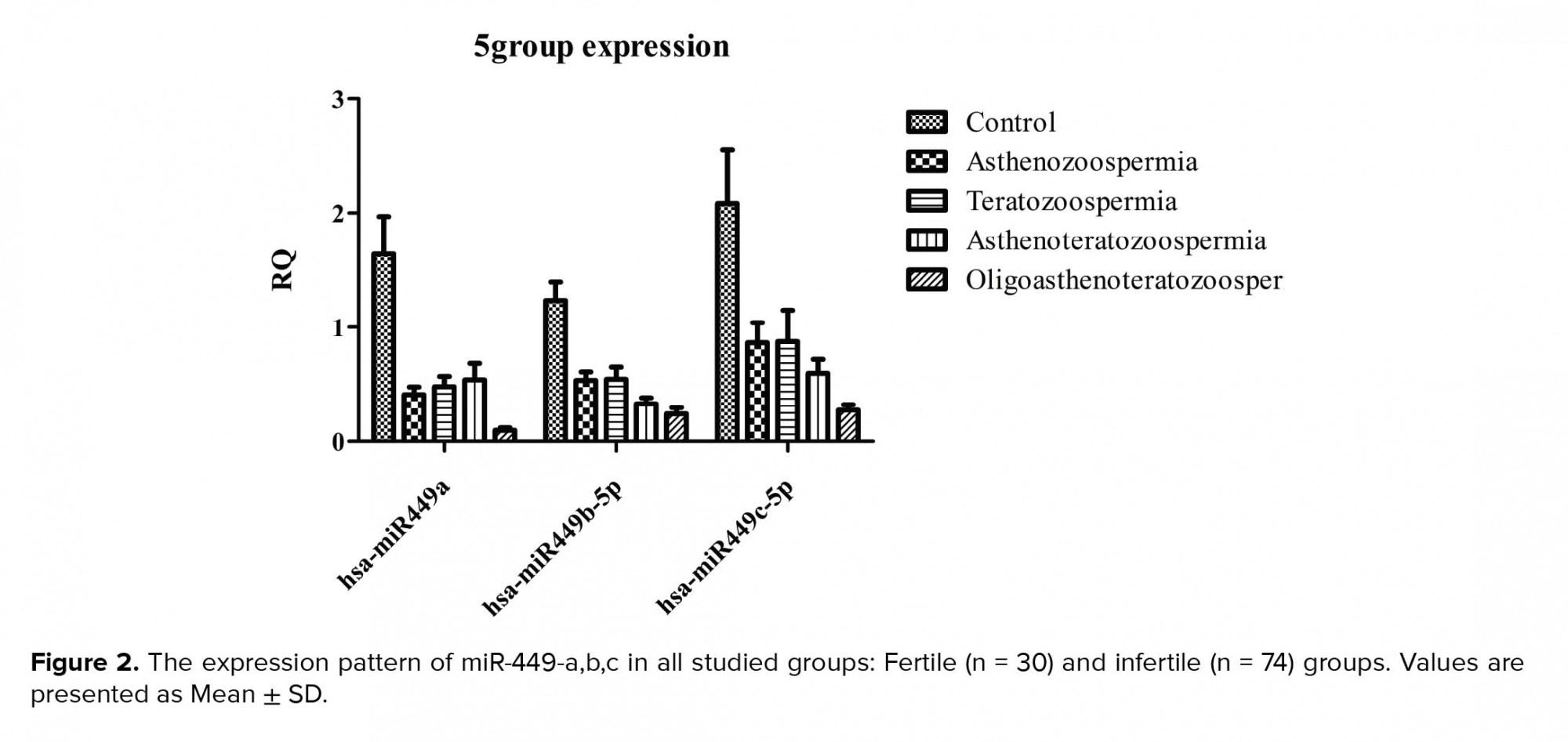
About the expression rate, we saw significant downregulation of the hsa-miR-449a expression in infertile group (0.24 ± 0.2) in comparison to the control groups (0.98 ± 0.37) (p = 0.0001). Also, significant down-egulation of this miRNA was shown in oligoasthenoterathospermia (0.05 ± 0.02), asthenospermia (0.24 ± 0.11), terathospermia (0.28 ± 0.2), and asthenoterathospermia (0.3 ± 0.11) as compared to the control group (0.98 ± 0.37) (F = 7.1 p = 0.0001). MirRNA expression ratio had not significant difference among the four infertile groups, (p = 0.21) (Figure 1A). The expression of hsa-miR-449b was downregulated significantly among the oligoasthenoterathospermia groups (0.022 ± 0.01) in comparison to the asthenospermia (0.04 ± 0.02), terathospermia (0.04 ± 0.032), asthenoterasthospermia (0.03 ± 0.02), and the control group (0.12 ± 0.09) (p = 0.0001, F = 2.9) (Figure 1B). However, in the case of hsa-miR-449c, oligoasthenoterathospermia patients showed significant downregulation of this miRNA (0.04 ± 0.03), also we observed this downregulation among the asthenospermia (0.15 ± 0.14), terathospermia (0.15 ± 0.14), and asthenoterasthospermia (0.1 ± 0.09) groups in compared to the control group )0.37 ± 0.01) (F = 5.04, p = 0.001) (Figure 1C). The average of the expression of the three studied miRNAs in fertile men showed that hsa-miR-449a had the highest levels of expression in these individuals )0.98 ± 0.3), followed by hsa-miR-449c (0.37 ± 0.01) and hsa-miR-449b (0.12 ± 0.09) (Figure 2). As displayed in Figure 2, among the three studied miRNAs, hsa-miR-449b had the lowest expression ratio in infertile men especially in oligoasthenotaratospermic men (0.022 ± 0.01).
3.2. Investigating the relationship between miRNAs expressions and parameters of sperm
In all studied samples, we observed significant correlation between the expression of hsa-miR-449a and the sperm progressive motility (r = 0.44, p = 0.0001), sperm count (r = 0.2, p = 0.03), and normal morphology (r = 0.46, p = 0.0001). The pearson test results showed, significant correlation between the expression of hsa-miR-449b miRNA and the spem progressive motility (r = 0.55, p = 0.0001), sperm count (r = 0.59, p = 0.0001), and normal morphology (r = 0.58, p = 0.0001). Also, there was significant correlation between the sperm count (r = 0.38, p = 0.0001), progressive motility (r = 0.22, p = 0.0322), and normal morphology (r = 0.32, p = 0.0001) with hsa-miR-449c experssion among all studied samples (Figure 3 A-I).
3.3. The methylation patterns of the promoter region of the hsa-miR-449a,b,c
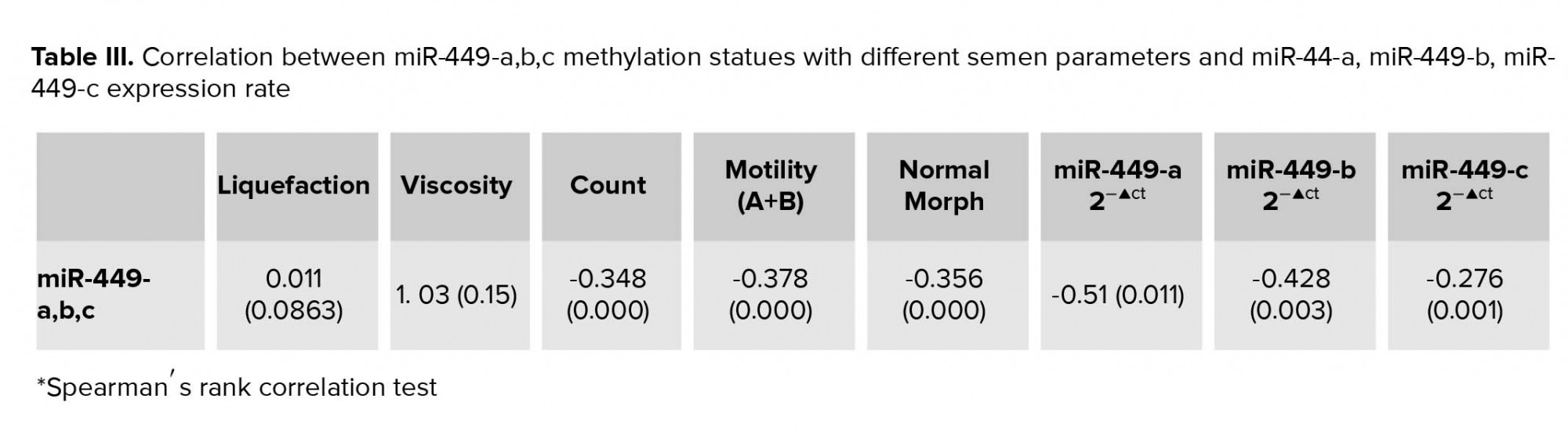
MSP results showed that, 60.8% and of the patients and 23.3% of the controls had methylated allele (p = 0.0001). The unmethylated allele was detected in all patients and controls. Oligoasthenoteratospermic (81.2%) and asthenothratospermic (61.2%) patients showed the highest frequency of methylation (Table II) (Figure 4 A, B). There was not significant correlation between hsa-hsa-miR-449abc promoter methylation with the liquefaction time (r = 0.022, p = 0.065) and viscosity (r = 0.969, p = 0.65). However, we observed a significant negative correlation between sperm count (r = -0.235, p = 0.003), progressive motility (r = -0.375, p = 0.0001), and normal morphology of sperms (r = -0.356, p = 0.0001) with methylation pattern of hsa-miR-449abc promoter. Also, there was significant negative correlation between methylation of hsa-miR-449abc promoter region and hsa-miR-449a (r = -0.110, p = 0.02), hsa-miR-449b (r = -0.245, p = 0.01), and hsa-miR-449c (r = -0.348, p = 0.005) expression ratio (Table III). The results of the effect of smoking on methylation status of mir-449-abc promoter showed the high frequency of methylation in men who smoked (87.7%) in comparison to men who did not (75.2%, χ2 = 4.2, p = 0.003).



4. Discussion
In our project, significant downregulation of the hsa-miR-449abc was seen in the sperm samples of infertile men. Briefly, miRNAs have the main role in incidence of male infertility through the gene expersion regulation (22). The miR-449 family have three members in mice and humans, hsa-miR-449a, hsa-miR-449b, and hsa-miR-449c. Three hsa-miR-499 (miR-499a, b, c) members are transcribed simultaneously and have the same seed sequence (17, 20).
The roles of hsa-miR-449 family in human reproduction, so far it has only been based on the results of animal models studeis (21). These studies report that hsa-miR-449 family is one of the most upregulated testicular miRNA and have major role in the initiation of meiotic phase in the adult testes. Also, this group showed that “hsa-miR-449 is predominantly and exclusively expressed in spermatocytes and spermatids in the adult testes” (21). Liu and colleague showed a presence of hsa-miR-449 family in spermatozoa, but their absence in oocytes (28). In the line of above study, our results showed expression of these miRNAs was shown in sperm,s s of the control (fertile) group, and hsa-miR-449a had the highest levels of expression. We also saw a significant downregulation of these miRNAs in the sperm samples of infertile patients, and hsa-miR-449b had the highest reduction. Another study reported abnormal different sperm parameters in hsa-miR-34bc/449 knock-out mice with oligoteratoasthenospermia. They reported that deficient mice had spermatozoa maturation stages problme, but hsa-miR-449 or hsa-miR-34 cluster Knockout (KO) mice showed no explained phenotype (22). This is because hsa-miR-449 and hsa-miR-34b/c have the same “seed sequence,” which maps between the second and seventh nucleotides; this core element is necessary for base pairing with target mRNAs, and can target the same set of mRNAs. On the other hand, hsa-miR-449 and hsa-miR-34b/c have the same expression profiles within testicular formation, and they are located to the precise identical spermatogenic cell types, such as spermatocytes and spermatids. In this regard, it is better in future studies that the expression pattern of hsa-miR-449 family and hsa-miR-34b/c be evaluated simultaneously. Interestingly, along with Comazzetto and colleague’s 2014 study, the highest downregulation of hsa-miR-449 family in oligoteratoasthenospermic men was observed, also we saw significant correlation between expression of this gene family and sperm progressive motility, count, and normal morphology (29).
Methylation is one of the mechanisms in expression regulation of miRNAs (30). Expression and methylation of the hsa-miR-449a,b,c as the tumor suppressor genes has been reported in different cancers such as prostate (31), hepatocellular carcinoma (32), and osteosarcoma (18). Also, the role of methylation in regulating the expression of miRNAs involved in spermatogenesis and infertility has also been demonstrated (33). To the best of our knowledge, there is no clinical data on the methylation status of hsa-miR-449a, b, c promoter in human sperm. In the study of hsa-miR-449family methylation, our results showed that, there was high frequency of methylation in infertile men (60.8%) specially in the oligoasthenoteratospermia patients. Interestingly, oligoasthenoteratospermia patients had the highest gene expression reduction. Also, in this research we observed a negative correlation between underexperssion and hypermethylation of hsa-miR-449a, b, c. This suggests that hypermethylation can be one of the downregulation mechanisms of these gene family. Further, we observed hypermethylation of promoter region among the 87.7% of smokers. Smoking and obesity have the major effect on the quantity of sperm miRNA (34). Recently, in the sperm samples of men exposed to lifestyle stress Dickson et al. reported under expression and hyper methylation of hsa-miR-449 and hsa-miR-34 (35). Acording to what was said, it is better to study the effect of other environmental factors on the rate of the hsa-miR-449a, b, c methylation.
5. Conclusion
Results of this research showed under-experssion and hypermethylation of the hsa-miR-449 a,b,c in sperm samples of infertile men. According to the results of this study, it can be said that one of the possible causes of defective spermatogenesis in infertile people can be reduced expression and increased methylation of these miRNAs family.
Acknowledgments
We appreciate all of the participants in current study. We also thank Cellular and Molecular Research Center of Qazvin University of Medical Science for providing laboratory facilities.
Conflict of Interest
The authors declare that there is no conflict of interest.
Full-Text: (410 Views)
1. Introduction
On average, it is estimated that infertility occurs in 10 to 15% of couples and 50% of infertility cases are due to male factor. The causes of 65-70% of male infertility cases is unknown and the correct mechanism has not been defined (1, 2). Genetic factors can be one of the causes of male infertility (3). But the role of miRNAs in the process of spermatogenesis and male infertility is very important. MiRNAs are non-coding RNAs, and play an important role in regulating gene expression (4, 5). MiRNAs regulate gene expression in two ways: by suppressing transcription and translation (RNAi) (6) or by activating transcription (RNAa) (7-9). The expression of miRNAs was shown in some types of male germ cells (10-14). The hsa-miR-449 was first detected in the fetal brain of mice(15, 16).
The hsa-miR-449 family has three members in mice and human, namely hsa-miR-449a, hsa-miR-449b, and hsa-miR-449c. These miRNAs have conserved sequence among different species and are located on the second intron of the Cdc20b gene. While the three hsa-miR-499 (miR-499 a, b, c) members have the same seed sequences, hsa-miR-449 members play the main role in the control of cell cycle and differentiation of epidermis (17-19).
In this regard, studies have shown that the hsa-miR449 family (hsa-miR-449a, hsa-miR449b, and hsa-miR-449c) and the hsa-miR-34 b, c (hsa-miR-34b-3p and hsa-miR-34c-5p) contain an identical seed sequence and have same sequence with the another miRNA, hsa-miR-34a-5p (17-21). About in addition, with respect to the role of hsa-miR-449 in spermatogenesis and male infertility, it was shown that the hsa-miR-449 family highly express in spermatocyte, spermatid, and adult testis. In one study was reported that, inactivation of both the hsa-miR34-b,c and hsa-miR-449 causes low sperm counts, motility and high abnormal sperm morphology in animal models (22). CpG methylation of mentioned genes is one possible reason for the their underexpression (20, 22), so that, under expression and high methylation of hsa-miR-449 (a, b, c) (as an important tumor-suppressor gene) have been shown in various cancers (23, 24). Besides, methylation of their promoter region has also been shown to be one of the mechanisms of expression reduction in adition to playing an important role in carcinogenesis of these microRNAs. Up to now, the patterns of hsa-miR-449 family expression and methylation has not been reported in different groups of infertile men and all the information in this regard has been based on animal models (25). Therefore, in this study we evaluated the expression and methylation pattern of hsa-miR-449 family in infertile men.
2. Materials and Methods
2.1. Subject recruitment and sampling
In this case-control study, 74 infertile men with idiopathic asthenozoospermia (n = 14), teratozoospermia (n = 16), asthenoteratozoospermia (n = 28), and oligoasthenoteratozoospermia (n = 16) were collected during 2018-2019 from the ACECR Telemedicine Infertility Center Qazvin, IRAN, based on the WHO criteria. The condition of infertile men was as follows: a history of infertility for at least 1 yr with their wives having a normal gynecological evaluation. However, infertile men conditions such as cystic fibrosis, Klinefelter syndrome, varicocele, chemotherapy, AZF, and genes micro deletions were not included in this study. In addition, 30 fertile healthy men were recruited as the control group. A questionnaire was designed to evaluation of the patients and controls’ information, including medical history, occupational and environmental condition, smoking condition (an adult who has smoked 100 cigarettes in his lifetime and who currently smokes cigarettes)and reproduction status (Table I). Patients were advised not to have sexual abstinence for three days before sampling. After sampling, semen samples were stored at 37ºC for 30 min to complete the liquefaction. Then, based on WHO criteria sperms concentration, motility, and morphology were evaluated (26).
2.2. RNA extraction and qRT-PCR
After liqufication, we centrifuged the semen samples for 10 min at 500 g. Then 1 ml FSB (Merck, Germany) was added to sperms pellet and somatic cells removed.TRIZOL reagent was used for isolation of the total RNA from the sperms based on kit protocols (Invitrogen Life Technology Co., USA). hsa-miR-449-a (MI0001648), hsa-miR-449-b (MI0003673), and hsa-449-c (MI0003823) were studied in this research. hsa-miR30a-5p (MIMAT0000087) and hsa-miR100-5p (MIMAT 0000102) were used as internal controls. Rotor gene-Q real-time PCR system (Qiagene, Germany) was used to quantifing of the RNA expression. Total amount of master mix was 10 µl and included 1 µl of reverse and forward primers (Exiqon, Denmark), 5 µl of Ampliqon real Q plus 2× master mix green (Ampliqone, Denmark), and 4 µl of diluted cDNA. For enzyme activation we incubated master mix for 15 min at 95ºC. Then reaction was runned in 40 cycle for 20 sec at 95ºC and 60 sec at 60ºC. Ct values was used for evaluation of expression rate of studied miRNAs. hsa-miR30a-5p and hsa-miR100-5p were used as the endogenous controls. The 2-△Ct method used for expression rate detection of target genes in comparision to internal controls.
2.3. DNA extraction and bisulfite modification
Phenol-chloroform method was used for DNA extraction. 2-5 µg of extracted DNA was bisulfited by using of EpiJET™ Bisulfite Conversion kit (Thermo Fisher Scientific, Inc).
2.4. MSPCR
The hsa-miR-449 methylation status was evaluated in all studied samples. For the targeted site, methylation-specific primers were designed. While the methylated primers were Forward: 5’-CGTTCGTTAATTTTTTCGTTTTTTGTCGC-3’) and Reverse: 5ꞌ-GTCAAAACCCGAATAAAATTCCCCGACG-3’, the unmethylated primers were Forward: 5’-TTGTTTGTTAATTTTTTTGTTTTTTGTTGT-3’ and Reverse: 5’- ATCAAAACCCAAATAAAATTCCCCAACA-3’. Methylation-specific PCR (MSP) was used to evaluation of methylation status of hsa-miR-449-abc promoter region. 1 µL of bisulfit converted DNA with methylated and unmethylated primers was amplifiedin in final 10 µL reaction mixture. "The PCR conditions for methylation status were: 95ºC for 15 min (Hot start), followed by 35 cycles at 95ºC for 20 sec (denaturation), 56.5ºC for 45 sec (annealing), and 72ºC for 45 sec (extension). PCR condition for unmethylated- primers was the same as the methylated condition except for the number of cycles and annealing temperature (60ºC)" (27).

2.5. Ethical considerations
This case-control study was approved by the Ethics Committee of Qazvin university of medical science with dedicated ID IR.QUMS.REC.1396.294. The recruited patients gave their informed written consent.
2.6. Statistical analysis
The GraphPad software (GraphPad PRISM V 5.04) was used for the data analysis. The analysis of variance test (ANOVA) was used for the evaluation of the miRNAs expression levels difference among the different studied groups. The frequency of promoter methylation pattern was evaluated by a nonparametric test (Kruskal-Wallis). The correlation between the miRNA expression rate, methylation with different sperm parameters was analyzed by Spearman's rank correlation. All P-values were two-tailed, with p < 0.05 considered as statistically significant.
3. Results
3.1. Expression of miRNAs in sperm samples of studied groups
About the expression rate, we saw significant downregulation of the hsa-miR-449a expression in infertile group (0.24 ± 0.2) in comparison to the control groups (0.98 ± 0.37) (p = 0.0001). Also, significant down-egulation of this miRNA was shown in oligoasthenoterathospermia (0.05 ± 0.02), asthenospermia (0.24 ± 0.11), terathospermia (0.28 ± 0.2), and asthenoterathospermia (0.3 ± 0.11) as compared to the control group (0.98 ± 0.37) (F = 7.1 p = 0.0001). MirRNA expression ratio had not significant difference among the four infertile groups, (p = 0.21) (Figure 1A). The expression of hsa-miR-449b was downregulated significantly among the oligoasthenoterathospermia groups (0.022 ± 0.01) in comparison to the asthenospermia (0.04 ± 0.02), terathospermia (0.04 ± 0.032), asthenoterasthospermia (0.03 ± 0.02), and the control group (0.12 ± 0.09) (p = 0.0001, F = 2.9) (Figure 1B). However, in the case of hsa-miR-449c, oligoasthenoterathospermia patients showed significant downregulation of this miRNA (0.04 ± 0.03), also we observed this downregulation among the asthenospermia (0.15 ± 0.14), terathospermia (0.15 ± 0.14), and asthenoterasthospermia (0.1 ± 0.09) groups in compared to the control group )0.37 ± 0.01) (F = 5.04, p = 0.001) (Figure 1C). The average of the expression of the three studied miRNAs in fertile men showed that hsa-miR-449a had the highest levels of expression in these individuals )0.98 ± 0.3), followed by hsa-miR-449c (0.37 ± 0.01) and hsa-miR-449b (0.12 ± 0.09) (Figure 2). As displayed in Figure 2, among the three studied miRNAs, hsa-miR-449b had the lowest expression ratio in infertile men especially in oligoasthenotaratospermic men (0.022 ± 0.01).
3.2. Investigating the relationship between miRNAs expressions and parameters of sperm
In all studied samples, we observed significant correlation between the expression of hsa-miR-449a and the sperm progressive motility (r = 0.44, p = 0.0001), sperm count (r = 0.2, p = 0.03), and normal morphology (r = 0.46, p = 0.0001). The pearson test results showed, significant correlation between the expression of hsa-miR-449b miRNA and the spem progressive motility (r = 0.55, p = 0.0001), sperm count (r = 0.59, p = 0.0001), and normal morphology (r = 0.58, p = 0.0001). Also, there was significant correlation between the sperm count (r = 0.38, p = 0.0001), progressive motility (r = 0.22, p = 0.0322), and normal morphology (r = 0.32, p = 0.0001) with hsa-miR-449c experssion among all studied samples (Figure 3 A-I).
3.3. The methylation patterns of the promoter region of the hsa-miR-449a,b,c
MSP results showed that, 60.8% and of the patients and 23.3% of the controls had methylated allele (p = 0.0001). The unmethylated allele was detected in all patients and controls. Oligoasthenoteratospermic (81.2%) and asthenothratospermic (61.2%) patients showed the highest frequency of methylation (Table II) (Figure 4 A, B). There was not significant correlation between hsa-hsa-miR-449abc promoter methylation with the liquefaction time (r = 0.022, p = 0.065) and viscosity (r = 0.969, p = 0.65). However, we observed a significant negative correlation between sperm count (r = -0.235, p = 0.003), progressive motility (r = -0.375, p = 0.0001), and normal morphology of sperms (r = -0.356, p = 0.0001) with methylation pattern of hsa-miR-449abc promoter. Also, there was significant negative correlation between methylation of hsa-miR-449abc promoter region and hsa-miR-449a (r = -0.110, p = 0.02), hsa-miR-449b (r = -0.245, p = 0.01), and hsa-miR-449c (r = -0.348, p = 0.005) expression ratio (Table III). The results of the effect of smoking on methylation status of mir-449-abc promoter showed the high frequency of methylation in men who smoked (87.7%) in comparison to men who did not (75.2%, χ2 = 4.2, p = 0.003).






4. Discussion
In our project, significant downregulation of the hsa-miR-449abc was seen in the sperm samples of infertile men. Briefly, miRNAs have the main role in incidence of male infertility through the gene expersion regulation (22). The miR-449 family have three members in mice and humans, hsa-miR-449a, hsa-miR-449b, and hsa-miR-449c. Three hsa-miR-499 (miR-499a, b, c) members are transcribed simultaneously and have the same seed sequence (17, 20).
The roles of hsa-miR-449 family in human reproduction, so far it has only been based on the results of animal models studeis (21). These studies report that hsa-miR-449 family is one of the most upregulated testicular miRNA and have major role in the initiation of meiotic phase in the adult testes. Also, this group showed that “hsa-miR-449 is predominantly and exclusively expressed in spermatocytes and spermatids in the adult testes” (21). Liu and colleague showed a presence of hsa-miR-449 family in spermatozoa, but their absence in oocytes (28). In the line of above study, our results showed expression of these miRNAs was shown in sperm,s s of the control (fertile) group, and hsa-miR-449a had the highest levels of expression. We also saw a significant downregulation of these miRNAs in the sperm samples of infertile patients, and hsa-miR-449b had the highest reduction. Another study reported abnormal different sperm parameters in hsa-miR-34bc/449 knock-out mice with oligoteratoasthenospermia. They reported that deficient mice had spermatozoa maturation stages problme, but hsa-miR-449 or hsa-miR-34 cluster Knockout (KO) mice showed no explained phenotype (22). This is because hsa-miR-449 and hsa-miR-34b/c have the same “seed sequence,” which maps between the second and seventh nucleotides; this core element is necessary for base pairing with target mRNAs, and can target the same set of mRNAs. On the other hand, hsa-miR-449 and hsa-miR-34b/c have the same expression profiles within testicular formation, and they are located to the precise identical spermatogenic cell types, such as spermatocytes and spermatids. In this regard, it is better in future studies that the expression pattern of hsa-miR-449 family and hsa-miR-34b/c be evaluated simultaneously. Interestingly, along with Comazzetto and colleague’s 2014 study, the highest downregulation of hsa-miR-449 family in oligoteratoasthenospermic men was observed, also we saw significant correlation between expression of this gene family and sperm progressive motility, count, and normal morphology (29).
Methylation is one of the mechanisms in expression regulation of miRNAs (30). Expression and methylation of the hsa-miR-449a,b,c as the tumor suppressor genes has been reported in different cancers such as prostate (31), hepatocellular carcinoma (32), and osteosarcoma (18). Also, the role of methylation in regulating the expression of miRNAs involved in spermatogenesis and infertility has also been demonstrated (33). To the best of our knowledge, there is no clinical data on the methylation status of hsa-miR-449a, b, c promoter in human sperm. In the study of hsa-miR-449family methylation, our results showed that, there was high frequency of methylation in infertile men (60.8%) specially in the oligoasthenoteratospermia patients. Interestingly, oligoasthenoteratospermia patients had the highest gene expression reduction. Also, in this research we observed a negative correlation between underexperssion and hypermethylation of hsa-miR-449a, b, c. This suggests that hypermethylation can be one of the downregulation mechanisms of these gene family. Further, we observed hypermethylation of promoter region among the 87.7% of smokers. Smoking and obesity have the major effect on the quantity of sperm miRNA (34). Recently, in the sperm samples of men exposed to lifestyle stress Dickson et al. reported under expression and hyper methylation of hsa-miR-449 and hsa-miR-34 (35). Acording to what was said, it is better to study the effect of other environmental factors on the rate of the hsa-miR-449a, b, c methylation.
5. Conclusion
Results of this research showed under-experssion and hypermethylation of the hsa-miR-449 a,b,c in sperm samples of infertile men. According to the results of this study, it can be said that one of the possible causes of defective spermatogenesis in infertile people can be reduced expression and increased methylation of these miRNAs family.
Acknowledgments
We appreciate all of the participants in current study. We also thank Cellular and Molecular Research Center of Qazvin University of Medical Science for providing laboratory facilities.
Conflict of Interest
The authors declare that there is no conflict of interest.
Type of Study: Original Article |
Subject:
Reproductive Genetics
References
1. Krausz C, Riera-Escamilla A. Genetics of male infertility. Nature Reviews Urology 2018; 15: 369-384. [DOI:10.1038/s41585-018-0003-3]
2. Freitas e Silva KS. Molecular genetics of male infertility: A mini-review. Trends in Res 2018; 1: 1-3. [DOI:10.15761/TR.1000112]
3. Kim SY, Kim HJ, Lee BY, Park SY, Lee HS, Seo JT. Y Chromosome microdeletions in infertile men with non-obstructive azoospermia and severe oligozoospermia. J Reprod Infert 2017; 18: 307-315.
4. O'Brien J, Hayder H, Zayed Y, Peng Ch. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology 2018; 9: 402-413. [DOI:10.3389/fendo.2018.00402]
5. Plotnikova O, Baranova A, Skoblov M. Comprehensive analysis of human microRNA-mRNA interactome. Frontiers in Genetics 2019; 10: 933-401. [DOI:10.3389/fgene.2019.00933]
6. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clinical Epigenetics 2019; 11: 25-48. [DOI:10.1186/s13148-018-0587-8]
7. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology 2019; 20: 21-37. [DOI:10.1038/s41580-018-0045-7]
8. Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RHA, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005; 120: 21-24. [DOI:10.1016/j.cell.2004.12.031]
9. Pasquinelli AE, Hunter Sh, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev 2005; 15: 200-205. [DOI:10.1016/j.gde.2005.01.002]
10. Kotaja N. Micro RNA and spermatogenesis. Fertility and Sterility 2014; 101: 1552-1562. [DOI:10.1016/j.fertnstert.2014.04.025]
11. Harton GL, Tempest HG. Chromosomal disorders and male infertility. Asian J Androl 2012; 14: 32-39. [DOI:10.1038/aja.2011.66]
12. Urdinguio RG, Bayon GF, Dmitrijeva M, Torano EG, Bravo C, Fraga MF, et al. Aberrant DNA methylation patterns of spermatozoa in men with nexplained infertility. Hum Reprod 2015; 30: 1014-1028. [DOI:10.1093/humrep/dev053]
13. Tuttelmann F, Simoni M, Kliesch S, Ledig S, Dworniczak B, Wieacker P, et al. Copy number variants in patients with severe oligozoospermia and Sertoli-cell-only syndrome. PLoS One 2011; 6: e19426. [DOI:10.1371/journal.pone.0019426]
14. Gunes S, Asci R, Okten G, Atac F, Onar OE, Ogur G, et al. Two males with SRY-positive 46, XX testicular disorder of sex development. Syst Biol Reprod Med 2013; 59: 42-47. [DOI:10.3109/19396368.2012.731624]
15. Pomper N, Liu Y, Hoye ML, Dougherty JD. CNS MicroRNA profiles: a database for cell type enriched microRNA expression across the mouse central nervous system. Sci Rep 2020; 20: 4914-4921. [DOI:10.1038/s41598-020-61307-5]
16. Zhang Zh, Zhuang L, Lin ChH. Roles of microRNAs in establishing and modulating stem cell potential. Int J Mol Sci 2019; 20: 3643-3673. [DOI:10.3390/ijms20153643]
17. Xie K, Liu J, Chen J, Dong J, Ma H, Liu Y, et al. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene 2014; 543: 101-107. [DOI:10.1016/j.gene.2014.03.059]
18. Yang X, Feng M, Jiang X, Wu Zh, Li Zh, Aau M, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev 2009; 23: 2388-2393. [DOI:10.1101/gad.1819009]
19. Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 2011; 12: 19-31. [DOI:10.1038/nrg2916]
20. Boeva V. Analysis of genomic sequence motifs for deciphering transcription factor binding and transcriptional regulation in eukaryotic cells. Front Genet 2016; 7: 1-16. [DOI:10.3389/fgene.2016.00024]
21. Bao J, Li D, Wang L, Wu J, Hu Y, Wang Zh, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem 2012; 287: 21686-21698. [DOI:10.1074/jbc.M111.328054]
22. Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, et al. The RNase III enzyme DROSHA is essential for MicroRNA production and spermatogenesis. J Biol Chem 2012; 287: 25173-25190. [DOI:10.1074/jbc.M112.362053]
23. Li Q, LI H, Zhao X, Wang B, Zhang L, Zhang C, et al. DNA methylation mediated downregulation of miR-449c controls osteosarcoma cell cycle progression by directly targeting oncogene c-Myc. Int J Biol Sci 2017; 13: 1038-1050. [DOI:10.7150/ijbs.19476]
24. Zhang Q, Yang Zh, Shan J, Liu L, Liu Ch, Shen J, et al. MicroRNA-449a maintains self-renewal in liver cancer stem-like cells by targeting Tcf3. Oncotarget 2017; 8: 110187-110200. [DOI:10.18632/oncotarget.22705]
25. Wu J, Bao J, Kim M, Yuan Sh, Tang Ch, Zheng H, et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci USA 2014; 111: E2851-E2857. [DOI:10.1073/pnas.1407777111]
26. World Health Organization Do RHaR. WHO Laboratory Manual for the Examination and Processing of Human Semen. Geneva: WHO Press; 2013.
27. Momeni AM, Najafiour R, Hamta A, Jahani S, Moghbelinejad S. Experssion and methylation pattern of has_miR_34 family in sperm samples of infertile men. Reprod Sci 2020; 27: 301-308. [DOI:10.1007/s43032-019-00025-4]
28. Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci 2012; 109: 490-494. [DOI:10.1073/pnas.1110368109]
29. Comazzetto S, Di Giacomo M, Rasmussen KD, Much Ch, Azzi Ch, Perlas E, et al. Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet 2014; 10: e1004597: 1-11. [DOI:10.1371/journal.pgen.1004597]
30. Nissan T, Parker R. Computational analysis of miRNA-mediated repression of translation: Implications for models of translation initiation inhibition. RNA 2008; 14: 1480-1491. [DOI:10.1261/rna.1072808]
31. Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009; 28: 1714-1724. [DOI:10.1038/onc.2009.19]
32. Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 2012; 143: 811-820. [DOI:10.1053/j.gastro.2012.05.033]
33. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13: 484-492. [DOI:10.1038/nrg3230]
34. Marczylo EL, Amoako AA, Konje JC, Gant TW, Marczylo TH. Smoking induces differential miRNA expression in human spermatozoa: A potential transgenerational epigenetic concern? Epigenetics 2012; 7: 432-439. [DOI:10.4161/epi.19794]
35. Dickson DA, Paulus JK, Mensah V, Lem J, Saavedra-Rodriguez L, Gentry A, et al. Reduced levels of miRNAs 449 and 34 in sperm of mice and men exposed to early life stress. Translational Psychiatry 2018; 8: 101-110. [DOI:10.1038/s41398-018-0146-2]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





