Fri, Jul 4, 2025
[Archive]
Volume 20, Issue 5 (May 2022)
IJRM 2022, 20(5): 399-404 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Arefnia M, Motovali-Bashi M, Javadirad S, Norioun H. Overexpression of hsa-miR-30a-5p and non-obstructive azoospermia: A case-control study. IJRM 2022; 20 (5) :399-404
URL: http://ijrm.ir/article-1-2234-en.html
URL: http://ijrm.ir/article-1-2234-en.html
1- Department of Cell and Molecular Biology and Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran.
2- Department of Cell and Molecular Biology and Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran. ,mbashi@sci.ui.ac.ir
3- Medical Genetics Department, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran.
2- Department of Cell and Molecular Biology and Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran. ,
3- Medical Genetics Department, Institute of Medical Biotechnology, National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran.
Full-Text [PDF 433 kb]
(1214 Downloads)
| Abstract (HTML) (1625 Views)
2.2. Sample size
Based on the prevalence of azoospermia in the population (1%) and previous studies (14, 15) the size of the sample needed was determined to be 30. The sample size was calculated by the formula:
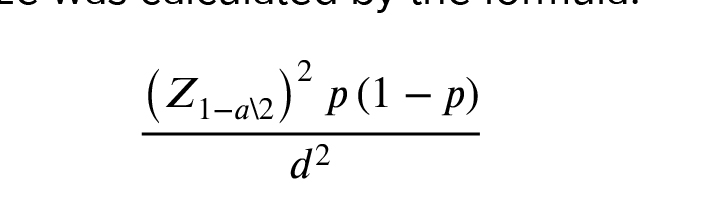
The current research was performed in 2020 on 30 azoospermic tissues. Samples were collected using testicular sperm extraction and microsurgical testicular sperm extraction from men with azoospermia who were referred to the Isfahan Fertility and Infertility Center (IFIC) in Isfahan, Iran from March 2013 to March 2015. Histology clarification suggested 11 samples as OA and 19 as NOA. All men who underwent the surgery had not had any previous medical history of testicular tumor or testis-related diseases. In accordance with the process used in previous studies on this topic, OA individuals were our control group (15-17).
In accordance with the manufacturer's instructions, 50 mg of testicular tissue was immersed in RNA later stabilization reagent (Ambion Life Science, Austin, TE, USA, AM7024). The testicular tissues were preserved at 4°C for 24 hr before being homogenized with a Heidolph Homogenizer DIAX 900 (Heidolph Instruments GmbH & CO. KG, Germany).
Sinaclon (Sinaclon, Iran) RNX Plus extraction buffer was used to extract total RNA. 1% agarose gel electrophoresis and Nanodrop OneC (Thermo Scientific, USA) were used to determine the quality and quantity of extracted RNA. RT-BON (Sinaclon, Iran) adaptor primer was used to add a poly (A) tail to the end of the extracted miRNAs, and oligo (dT) primers were used for the first-strand complementary DNA synthesis. Highly specific forward and reverse primers of hsa-miR-30a-5p were bought from Bonyakhteh Co, Iran (http://strc.ac.ir/en). The reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) was carried out in triplicate on a Choromo 4 Bio-Rad machine (USA). U6-snRNA was used as the calibrator gene (15).
2.3. Ethical considerations
Written informed consent was obtained from all individuals. The study protocol was approved by the institutional review board and Ethics Committee of Royan Institute, Isfahan, Iran (Code: IR.ACECR.ROYAN.REC.1394.105).
2.4. Statistical analysis
To calculate and compare the mean Cq of hsa-miR-30a-5p between men with OA vs. NO, t test were calculated using GraphPad Prism 7.05 (USA) software. P-values < 0.05 were considered significant.
Full-Text: (333 Views)
1. Introduction
Infertility is defined by the World Health Organization as an inability to conceive after at least 1 yr of regular unprotected intercourse (1). It is estimated that 15% of all couples in the world may experience infertility. Half of these failures are thought to have roots in male factors (2). Azoospermia is considered to be one of the main causes of male infertility, described as the lack of sperm in ejaculation. Azoospermia can result in infertility if an obstruction occurs in the seminiferous tubules; however, non-obstructive azoospermia (NOA) can develop independently of physical defects (3). Therefore, in people suffering from obstructive azoospermia (OA), the process of spermatogenesis is normal, but individuals with NOA show abnormal testicular spermatogenesis (4).
Animal knock-out models have supported the key role of the KDM3A gene for the proper development of spermatids. The main function of KDM3A is to demethylate H3K9s in the promoter region of the TNP1, PRM1, and PRM2 genes to facilitate the expression of these downstream genes (5). Taken together, the importance of the KDM3A gene to maintain the proper maturation of the male germ cells seems to be undeniable (6).
Micro-RNAs (miRNAs) are single-stranded evolutionarily conserved RNA molecules with 18-25 nucleotides in length. While these small RNAs are not translated in cells, they control the function of the cells via the degradation of the other target genes during their translation (7). Few investigations have demonstrated the roles of miRNAs in the development of male infertility (8, 9). The pioneer study (conducted in 2009) introduced mir-122a as a regulatory molecule of the Tnp2 gene (10). Other studies have highlighted the significance of miRNAs in spermatogenesis and the development of the masculine gametes (11-13).
Aberrant expression of KDM3A was reported previously, but the role of miRNAs controller was not discussed (5). To answer this question, hsa-miR-30a-5p expression in men with OA and NOA was investigated in this study.
Animal knock-out models have supported the key role of the KDM3A gene for the proper development of spermatids. The main function of KDM3A is to demethylate H3K9s in the promoter region of the TNP1, PRM1, and PRM2 genes to facilitate the expression of these downstream genes (5). Taken together, the importance of the KDM3A gene to maintain the proper maturation of the male germ cells seems to be undeniable (6).
Micro-RNAs (miRNAs) are single-stranded evolutionarily conserved RNA molecules with 18-25 nucleotides in length. While these small RNAs are not translated in cells, they control the function of the cells via the degradation of the other target genes during their translation (7). Few investigations have demonstrated the roles of miRNAs in the development of male infertility (8, 9). The pioneer study (conducted in 2009) introduced mir-122a as a regulatory molecule of the Tnp2 gene (10). Other studies have highlighted the significance of miRNAs in spermatogenesis and the development of the masculine gametes (11-13).
Aberrant expression of KDM3A was reported previously, but the role of miRNAs controller was not discussed (5). To answer this question, hsa-miR-30a-5p expression in men with OA and NOA was investigated in this study.
2. Materials and Methods
2.1. Bioinformatics investigation
To find the best candidate miRNA against KDM3A transcription, some online databases were explored:
To find the best candidate miRNA against KDM3A transcription, some online databases were explored:
- miRBase (http://www.mirbase.org),
- TargetScan (http://www.targetscan.org),
- DianamicroT (http://diana.imis.athenainnovation.gr),
- miRanda (http://www.microrna.org),
- mirwalk (http://zmf.umm.uniheidelberg.de),
- MirDB (http://www.mirdb.org),
- PicTar (https://pictar.mdcberlin.de),
- MiRGen (http://carolina.imis.athenainnovation.gr)
2.2. Sample size
Based on the prevalence of azoospermia in the population (1%) and previous studies (14, 15) the size of the sample needed was determined to be 30. The sample size was calculated by the formula:
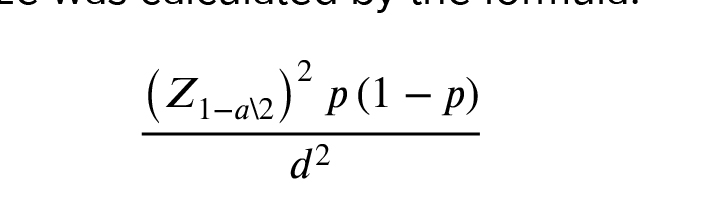
The current research was performed in 2020 on 30 azoospermic tissues. Samples were collected using testicular sperm extraction and microsurgical testicular sperm extraction from men with azoospermia who were referred to the Isfahan Fertility and Infertility Center (IFIC) in Isfahan, Iran from March 2013 to March 2015. Histology clarification suggested 11 samples as OA and 19 as NOA. All men who underwent the surgery had not had any previous medical history of testicular tumor or testis-related diseases. In accordance with the process used in previous studies on this topic, OA individuals were our control group (15-17).
In accordance with the manufacturer's instructions, 50 mg of testicular tissue was immersed in RNA later stabilization reagent (Ambion Life Science, Austin, TE, USA, AM7024). The testicular tissues were preserved at 4°C for 24 hr before being homogenized with a Heidolph Homogenizer DIAX 900 (Heidolph Instruments GmbH & CO. KG, Germany).
Sinaclon (Sinaclon, Iran) RNX Plus extraction buffer was used to extract total RNA. 1% agarose gel electrophoresis and Nanodrop OneC (Thermo Scientific, USA) were used to determine the quality and quantity of extracted RNA. RT-BON (Sinaclon, Iran) adaptor primer was used to add a poly (A) tail to the end of the extracted miRNAs, and oligo (dT) primers were used for the first-strand complementary DNA synthesis. Highly specific forward and reverse primers of hsa-miR-30a-5p were bought from Bonyakhteh Co, Iran (http://strc.ac.ir/en). The reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) was carried out in triplicate on a Choromo 4 Bio-Rad machine (USA). U6-snRNA was used as the calibrator gene (15).
2.3. Ethical considerations
Written informed consent was obtained from all individuals. The study protocol was approved by the institutional review board and Ethics Committee of Royan Institute, Isfahan, Iran (Code: IR.ACECR.ROYAN.REC.1394.105).
2.4. Statistical analysis
To calculate and compare the mean Cq of hsa-miR-30a-5p between men with OA vs. NO, t test were calculated using GraphPad Prism 7.05 (USA) software. P-values < 0.05 were considered significant.
3. Results
3.1. Bioinformatics exploration
Having explored various databases, we screened the existing information and hsa-miR-30a-5p was chosen. It was observed that hsa-miR-30a-5p showed the highest frequency among different databases with the highest score for the potential to impact KDM3A transcription.
3.2. RNA assessment
Agarose gel electrophoresis showed a 28S rRNA band intensity 2 times greater than 18S rRNA, an indication of the appropriate handling and intactness of the extracted RNAs (Figure 1). RNA absorbance ratios of 260/280 and 260/230 were between 1/8-2, and the purity of the extracted RNAs was confirmed.
3.3. RT-qPCR analysis
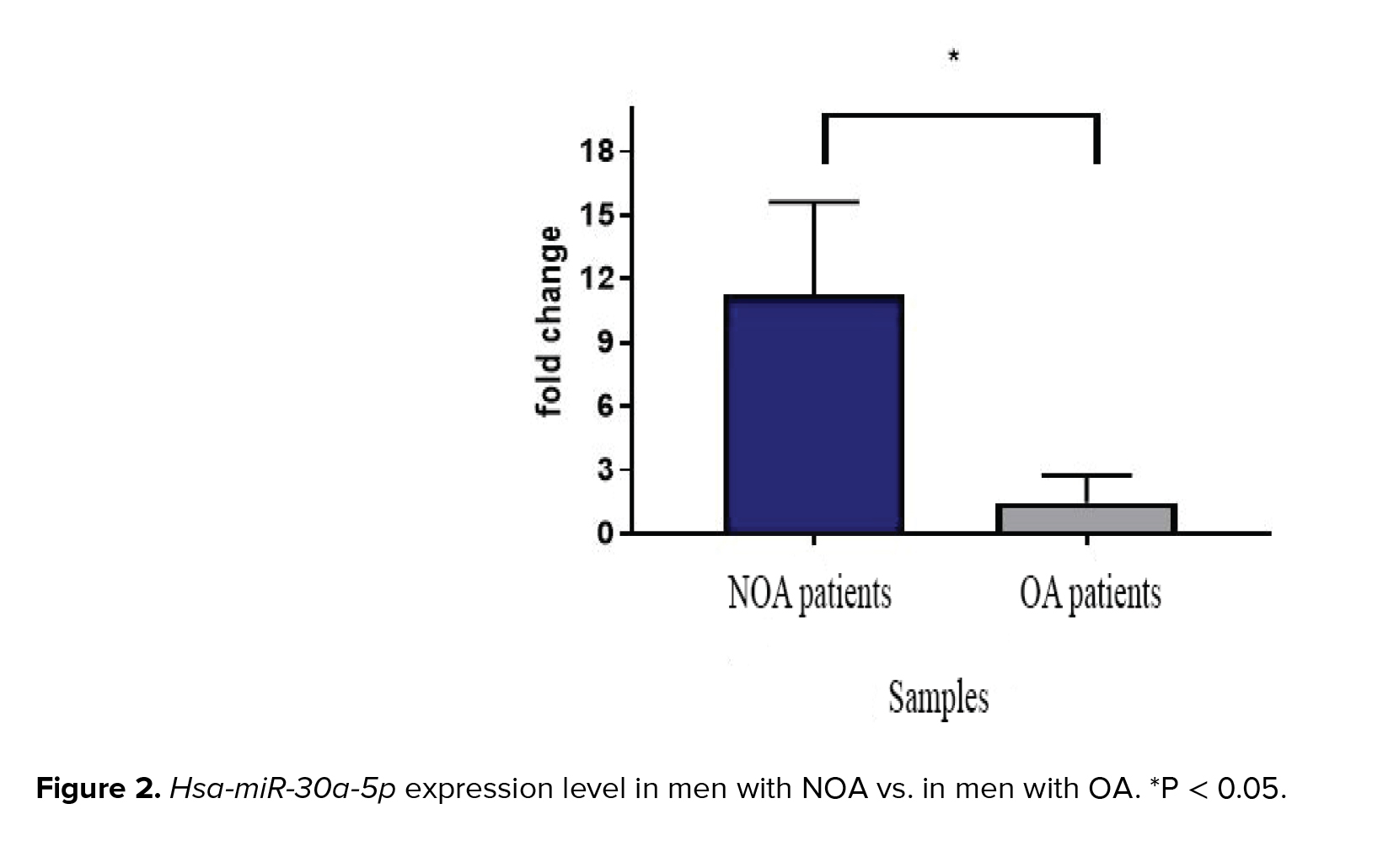
Melt curve analysis showed the highest specificity of the designed primer. The single pick of melt curve indicated the specificity of the primers and loss of primer dimer formation. The expression levels of hsa-miR-30a-5p in individuals with NOA and OA are illustrated in figure 2. Significant overexpression of hsa-miR-30a-5p was observed when men with NOA were compared to OA-control individuals (p = 0.04).

In contrast, another study showed the down-regulation of hsa-miR-30a-5p in men with NOA. The inconsistency might be ascribed to the control individuals as they used men with testicular carcinomas as their control group (18). In accordance with our results, hsa-miR-30a-5p was previously identified by microarray data analysis as a suggested biomarker in asthenozoospermia (19).
Among all the different cell populations in testicular tissue, the haploid cells and late meiotic cells are the fundamental sources of miRNA production during spermatogenesis (20). The first expression of miRNAs in testicular tissue was reported in 2004 (21). Subsequently, researchers focused on finding a function of miRNAs in the physiology of the germinal duct. In 2005, for the first time, it was found that mir-122a caused a negative expression regulation of the Tnp2 gene during spermatogenesis. It has also been shown that the function of this miRNA is to neutralize the expression of the gene through degradation of the gene’s transcription. Also, by using the luciferase assay technique, it was revealed that 15 picomoles of mir-122a can reduce the gene’s expression (22). A microarray data analysis revealed 154 down-regulated and 19 up-regulated miRNAs in testicular tissues and presented the first biomarkers of NOA (23). Another microarray analysis revealed 54 up-regulated and 27 down-regulated miRNAs in asthenozoospermia patients (19). Our investigation on hsa-miR-30a-5p suggested that overexpression of the mentioned miRNA leads to male infertility. Our study obtained similar results as another conducted on hsa-mir-27a-3p and its target KDM3A. The mentioned study revealed hsa-mir-27a-3p was overexpressed in NOA patients; it targeted the KDM3A transcript and consequently led to male infertility (15). Our study also suggested that overexpression of hsa-miR-30a-5p can lead to infertility in men with NOA.
Our data are contrary to a study that was conducted on prostate cancer. In that study microarray data indicated hsa-miR-30a-5p down-regulation occurred in NOA patients compared to prostate cancer patients. The difference in the results obtained may be due to differences in sample numbers and in the populations of the 2 studies. In that study, 3 individuals with NOA were sampled in China, but in the present study, 19 Iranian NOA patients were used for the study. Moreover, tissue heterogeneity may have led to a difference in the results of these 2 studies. The important point to be considered is the difference between the control groups of the studies. The mentioned study compared hsa-miR-30a-5p expression in individuals with NOA vs. in prostate cancer patients, but in the present study, the expression of miRNA in NOA patients was compared with that of OA patients (23). Hence, the results of both studies could be considered accurate.
The results of this study and a previous research show that hsa-miR-30a-5p can be considered as a biomarker for NOA and asthenozoospermia (19). Due to the network's expression and interconnected miRNAs, the findings can indicate a relationship between the expression of hsa-miR-30a-5p and the occurrence of NOA and asthenozoospermia diseases.
3.2. RNA assessment
Agarose gel electrophoresis showed a 28S rRNA band intensity 2 times greater than 18S rRNA, an indication of the appropriate handling and intactness of the extracted RNAs (Figure 1). RNA absorbance ratios of 260/280 and 260/230 were between 1/8-2, and the purity of the extracted RNAs was confirmed.
3.3. RT-qPCR analysis
Melt curve analysis showed the highest specificity of the designed primer. The single pick of melt curve indicated the specificity of the primers and loss of primer dimer formation. The expression levels of hsa-miR-30a-5p in individuals with NOA and OA are illustrated in figure 2. Significant overexpression of hsa-miR-30a-5p was observed when men with NOA were compared to OA-control individuals (p = 0.04).


4. Discussion
In the current study, 30 testicular tissues of idiopathic azoospermia from 19 individuals with NOA and 11 individuals with OA (control) were explored. RT-qPCR data analyses indicated that the expression of hsa-miR-30a-5p was 11.3 times higher in men with NOA compared to OA-control individuals (p = 0.04).In contrast, another study showed the down-regulation of hsa-miR-30a-5p in men with NOA. The inconsistency might be ascribed to the control individuals as they used men with testicular carcinomas as their control group (18). In accordance with our results, hsa-miR-30a-5p was previously identified by microarray data analysis as a suggested biomarker in asthenozoospermia (19).
Among all the different cell populations in testicular tissue, the haploid cells and late meiotic cells are the fundamental sources of miRNA production during spermatogenesis (20). The first expression of miRNAs in testicular tissue was reported in 2004 (21). Subsequently, researchers focused on finding a function of miRNAs in the physiology of the germinal duct. In 2005, for the first time, it was found that mir-122a caused a negative expression regulation of the Tnp2 gene during spermatogenesis. It has also been shown that the function of this miRNA is to neutralize the expression of the gene through degradation of the gene’s transcription. Also, by using the luciferase assay technique, it was revealed that 15 picomoles of mir-122a can reduce the gene’s expression (22). A microarray data analysis revealed 154 down-regulated and 19 up-regulated miRNAs in testicular tissues and presented the first biomarkers of NOA (23). Another microarray analysis revealed 54 up-regulated and 27 down-regulated miRNAs in asthenozoospermia patients (19). Our investigation on hsa-miR-30a-5p suggested that overexpression of the mentioned miRNA leads to male infertility. Our study obtained similar results as another conducted on hsa-mir-27a-3p and its target KDM3A. The mentioned study revealed hsa-mir-27a-3p was overexpressed in NOA patients; it targeted the KDM3A transcript and consequently led to male infertility (15). Our study also suggested that overexpression of hsa-miR-30a-5p can lead to infertility in men with NOA.
Our data are contrary to a study that was conducted on prostate cancer. In that study microarray data indicated hsa-miR-30a-5p down-regulation occurred in NOA patients compared to prostate cancer patients. The difference in the results obtained may be due to differences in sample numbers and in the populations of the 2 studies. In that study, 3 individuals with NOA were sampled in China, but in the present study, 19 Iranian NOA patients were used for the study. Moreover, tissue heterogeneity may have led to a difference in the results of these 2 studies. The important point to be considered is the difference between the control groups of the studies. The mentioned study compared hsa-miR-30a-5p expression in individuals with NOA vs. in prostate cancer patients, but in the present study, the expression of miRNA in NOA patients was compared with that of OA patients (23). Hence, the results of both studies could be considered accurate.
The results of this study and a previous research show that hsa-miR-30a-5p can be considered as a biomarker for NOA and asthenozoospermia (19). Due to the network's expression and interconnected miRNAs, the findings can indicate a relationship between the expression of hsa-miR-30a-5p and the occurrence of NOA and asthenozoospermia diseases.
5. Conclusion
According to the current study, hsa-miR-30a-5p is overexpressed in NOA patients compared to OA patients, and hsa-miR-30a-5p could potentially target the KDM3A transcript and hinder it from being translated. As a result, the downstream genes TNP1, PRM1, and PRM2 remain silent. All of these factors combine to induce male infertility in men with NOA.Acknowledgments
This study was performed and financially supported by the Graduate Studies Department of the University of Isfahan, Isfahan, Iran (Grant number: 12062).Conflict of Interest
The authors declare that there is no conflict of interest.
Type of Study: Original Article |
Subject:
Reproductive Genetics
References
1. Fernández Pelegrina R, Kessler AG, Rawlins RG. Modern approaches to the treatment of human infertility through assisted reproduction. P R Health Sci J 1991; 10: 75-81.
2. Irvine DS. Epidemiology and aetiology of male infertility. Hum Reprod 1998; 13 (Suppl.): 33-44. [DOI:10.1093/humrep/13.suppl_1.33] [PMID]
3. Aziz N. The importance of semen analysis in the context of azoospermia. Clinics 2013; 68 (Suppl.): 35-38. [DOI:10.6061/clinics/2013(Sup01)05]
4. Jarow JP, Espeland MA, Lipshultz LI. Evaluation of the azoospermic patient. J Urol 1989; 142: 62-65. [DOI:10.1016/S0022-5347(17)38662-7]
5. Javadirad SM, Hojati Z, Ghaedi K, Nasr‐Esfahani MH. Expression ratio of histone demethylase KDM 3A to protamine‐1 mRNA is predictive of successful testicular sperm extraction in men with obstructive and non‐obstructive azoospermia. Andrology 2016; 4: 492-499. [DOI:10.1111/andr.12164] [PMID]
6. Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450: 119-123. [DOI:10.1038/nature06236] [PMID]
7. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11: 597-610. [DOI:10.1038/nrg2843] [PMID]
8. Momeni A, Najafipour R, Hamta A, Jahani S, Moghbelinejad S. Expression and methylation pattern of hsa-miR-34 family in sperm samples of infertile men. Reprod Sci 2020; 27: 301-308. [DOI:10.1007/s43032-019-00025-4] [PMID]
9. Wang Ch, Yang C, Chen X, Yao B, Yang Ch, Zhu Ch, et al. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem 2011; 57: 1722-1731. [DOI:10.1373/clinchem.2011.169714] [PMID]
10. Liu T, Huang Y, Liu J, Zhao Y, Jiang L, Huang Q, et al. MicroRNA-122 influences the development of sperm abnormalities from human induced pluripotent stem cells by regulating TNP2 expression. Stem Cells Dev 2013; 22: 1839-1850. [DOI:10.1089/scd.2012.0653] [PMID] [PMCID]
11. Yan N, Lu Y, Sun H, Qiu W, Tao D, Liu Y, et al. Microarray profiling of microRNAs expressed in testis tissues of developing primates. J Assist Reprod Genet 2009; 26: 179-186. [DOI:10.1007/s10815-009-9305-y] [PMID] [PMCID]
12. Kotaja N. MicroRNAs and spermatogenesis. Fertil Steril 2014; 101: 1552-1562. [DOI:10.1016/j.fertnstert.2014.04.025] [PMID]
13. Walker WH. Regulation of mammalian spermatogenesis by miRNAs. Semin Cell Dev Biol 2021; S1084-9521(21)00118-X.
14. Charan J, Biswas T. How to calculate sample size for different study designs in medical research? Indian J Psychol Med 2013; 35: 121-126. [DOI:10.4103/0253-7176.116232] [PMID] [PMCID]
15. Norioun H, Motovali-Bashi M, Javadirad SM. Hsa-miR-27a-3p overexpression in men with nonobstructive azoospermia: A case-control study. Int J Reprod BioMed 2020; 18: 961-968. [DOI:10.18502/ijrm.v13i11.7963] [PMID] [PMCID]
16. Bonaparte E, Moretti M, Colpi GM, Nerva F, Contalbi G, Vaccalluzzo L, et al. ESX1 gene expression as a robust marker of residual spermatogenesis in azoospermic men. Hum Reprod 2010; 25: 1398-1403. [DOI:10.1093/humrep/deq074] [PMID]
17. Pansa A, Sirchia SM, Melis S, Giacchetta D, Castiglioni M, Colapietro P, et al. ESX1 mRNA expression in seminal fluid is an indicator of residual spermatogenesis in non-obstructive azoospermic men. Hum Reprod 2014; 29: 2620-2627. [DOI:10.1093/humrep/deu261] [PMID]
18. Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol 2009; 7: 13. [DOI:10.1186/1477-7827-7-13] [PMID] [PMCID]
19. Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, et al. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril 2013; 99: 1249-1255. [DOI:10.1016/j.fertnstert.2012.11.054] [PMID]
20. Abu‐Halima M, Galata V, Backes C, Keller A, Hammadeh M, Meese E. MicroRNA signature in spermatozoa and seminal plasma of proven fertile men and in testicular tissue of men with obstructive azoospermia. Andrologia 2020; 52: e13503. [DOI:10.1111/and.13503] [PMID]
21. Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res 2004; 14: 2486-2494. [DOI:10.1101/gr.2845604] [PMID] [PMCID]
22. Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod 2005; 73: 427-433. [DOI:10.1095/biolreprod.105.040998] [PMID]
23. Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol 2009; 7: 1-10. [DOI:10.1186/1477-7827-7-13] [PMID] [PMCID]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








