Sun, Dec 14, 2025
[Archive]
Volume 23, Issue 2 (February 2025)
IJRM 2025, 23(2): 111-130 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Keshari J R, Prakash P, Sinha S R, Prakash P, Rani K, Aziz T et al . Diagnostic potential of cell-free fetal nucleic acids in predicting pregnancy complications: A systematic review and meta-analysis on trisomy, pre-eclampsia, and gestational diabetes. IJRM 2025; 23 (2) :111-130
URL: http://ijrm.ir/article-1-3340-en.html
URL: http://ijrm.ir/article-1-3340-en.html
Jiut Ram Keshari *1 

 , Pritam Prakash2
, Pritam Prakash2 

 , Seema Rani Sinha2
, Seema Rani Sinha2 

 , Prem Prakash3
, Prem Prakash3 

 , Kirti Rani2
, Kirti Rani2 

 , Tarique Aziz4
, Tarique Aziz4 

 , Shaily Shilpa2
, Shaily Shilpa2 




 , Pritam Prakash2
, Pritam Prakash2 

 , Seema Rani Sinha2
, Seema Rani Sinha2 

 , Prem Prakash3
, Prem Prakash3 

 , Kirti Rani2
, Kirti Rani2 

 , Tarique Aziz4
, Tarique Aziz4 

 , Shaily Shilpa2
, Shaily Shilpa2 


1- Department of Biochemistry, Indira Gandhi Institute of Medical Sciences, Patna, Bihar, India. , jiutram@rediffmail.com
2- Department of Biochemistry, Indira Gandhi Institute of Medical Sciences, Patna, Bihar, India.
3- Department of General Surgery, Indira Gandhi Institute of Medical Sciences, Patna, Bihar, India.
4- Department of Biochemistry, Rajendra Institute of Medical Sciences, Ranchi, Jharkhand, India.
2- Department of Biochemistry, Indira Gandhi Institute of Medical Sciences, Patna, Bihar, India.
3- Department of General Surgery, Indira Gandhi Institute of Medical Sciences, Patna, Bihar, India.
4- Department of Biochemistry, Rajendra Institute of Medical Sciences, Ranchi, Jharkhand, India.
Full-Text [PDF 1700 kb]
(832 Downloads)
| Abstract (HTML) (835 Views)
Full-Text: (58 Views)
1. Introduction
In antenatal care, prenatal screening and diagnosis are routinely provided. It is believed that such amenities help women make decisions about whether to continue with pregnancies that are affected by developmental abnormalities. At present, the gold standard for prenatal diagnosis is the invasive method of acquiring fetal tissue and DNA. Though there is a little but considerable chance of miscarriage associated with invasive testing, many women prefer not to have it done (1, 2). To lower this risk of miscarriage and enable early testing, a reliable and feasible noninvasive prenatal diagnostic approach has long been sought upon.
The detection of cell-free fetal (cff) nucleic acids in the maternal bloodstream during pregnancy has completely changed the landscape of noninvasive prenatal screening and diagnosis. Circulating nucleic acids in maternal plasma and serum were anticipated to become a new area of interest in prenatal diagnostics following their discovery in 1997 (3). Although new in vivo studies show the possibility of other processes, cffDNA is released on trophoblastic cells as a result of apoptotic mechanisms. Since the rise in cffDNA levels occurs before the disease's clinical signs, it is highly valuable in the field of prenatal diagnosis and in predicting frequent pregnancy complications. Prenatal care has quickly included noninvasive prenatal screening utilizing cell-free DNA (cfDNA) in identifying common fetal aneuploidies, since its debut in 2011. According to a recent systematic review, the best sensitive and specific screening test for fetal Down syndrome, T13, and T18 in singleton and twin pregnancies is non-invasive prenatal screening with cfDNA. It was able to detect maternal diseases such as aneuploidies and malignancies, in contrast with conventional serum analyte screening (4).
While circulating cfDNA was the first known circulating nucleic acid, more have been discovered as molecular biology research and technology have advanced. At present, we take into consideration circulating nucleic acids in maternal plasma and serum, which is an array of extracellular nucleic acids that includes placental RNA, messenger RNA (mRNA), microRNA (miRNA), and other novel species like circular RNA, long noncoding RNA, mitochondrial DNA, and cfDNA (5). Alternative methods have been continuously explored, such as measuring total cfDNA or using gender-independent fetal epigenetic markers like DNA methylation and cfRNA of placental origin. These offer an appropriate replacement for genetic disease screening and prenatal diagnosis (6).
Recent research has demonstrated a correlation between elevated levels of cffDNA in maternal blood and various pregnancy complications, such as early pregnancy loss and preterm labor (7), pre-eclampsia (PE) (8-10), and fetal growth restriction (FGR) (11). Pregnancy difficulties have prompted studies to investigate the possibility of using cffDNA concentrations as a biomarker (12, 13). Research on the potential pathways connecting cffDNA to the etiology of pregnancy-related problems is only getting started (13).
We conducted a comprehensive review and meta-analysis to determine if the cff nucleic acid identification markers in the maternal bloodstream may be identified as diagnostic risk factors for fetal outcomes.
2. Materials and Methods
2.1. Search strategy
A comprehensive literature search was conducted in 7 different databases (PubMed, The Cochrane Library, Embase, Science Direct, Web of Science, Scopus, and Google Scholar). Preferred reporting items for systematic review and meta-analyses (PRISMA) checklist guidelines were followed (14). Articles published between 2010 and October 2023 were included in the study. During the literature search, restrictions were not placed on country, time, or language of publication. Editorial letters, conference proceedings, and practice guidelines were excluded.
The following key terms were used to identify relevant studies: cffDNA, next-generation sequencing (NGS), ctDNA, fetal outcomes, fetal anomalies, cffDNA, cff nucleic acids, epigenetic methylation, noninvasive prenatal diagnosis, prenatal diagnosis, and genetic diagnosis. Only research articles were retrieved and reviewed. All possible combinations of keywords were utilized in the search. Table I shows the search strings used to retrieve research articles from each database. After removing duplicates, titles and abstracts were screened as per eligibility criteria. Full-text articles of all the identified abstracts were reviewed independently.
2.2. Criteria for considering studies
Inclusion criteria encompass published research (case-control, cross-sectional, prospective, and retrospective cohorts) examining cff nucleic acid markers in maternal circulation. Studies on adult women ( ≥ 18 yr) with pregnancy-related complications were only included. Exclusion criteria encompass studies lacking relevance or adequate data, non-English publications, and certain study types (e.g., reviews, meta-analyses, letters to editors, commentaries, guidelines, book chapters, and editorials). Both published and unpublished studies, including gray literature, were considered for inclusion. The review also screened reference lists of prior systematic reviews/meta-analyses. Details such as authors, year of publication, country, study design, sample size, cell-free nucleic acid involved, pregnancy-related complications, and important results relevant to the study objectives were extracted from the included studies. The meta-analysis included studies based on specific criteria: relevant study design, availability of comparable outcome measures, sufficient statistical data, methodological quality, similar populations and interventions, and peer-reviewed full-text publications.
2.3. Study quality assessment
The Joanna Briggs Institute's critical appraisal checklists for studies were used to assess the quality of selected studies (15). A study was deemed to have a high risk of bias if the "yes" score was 49% or lower, moderate risk if the score was between 50 and 69%, and low risk if the score was 70% or higher. Each study underwent bias assessment and classification as low risk, high risk, or with some concerns. Any discrepancies between independent reviewers were resolved by consulting a third reviewer.
2.4. Statistical analysis and meta-analysis
For this meta-analysis, STATA version 17 was utilized. Continuous data were presented as means or medians with appropriate measures of variability. Categorical variables were expressed as numbers and percentages. A pooled meta-analysis was conducted. Heterogeneity was evaluated using Cochran’s Q test and the DerSimonian and Laird method. Heterogeneity levels were categorized as low, moderate, or high based on I-square values. Low heterogeneity was defined as an I-square value of < 25%, moderate heterogeneity as 25-50%, and high heterogeneity as > 50%. Publication bias was assessed visually with funnel plots and quantitatively using Egger’s test.


3. Results
3.1. Identification and description of studies
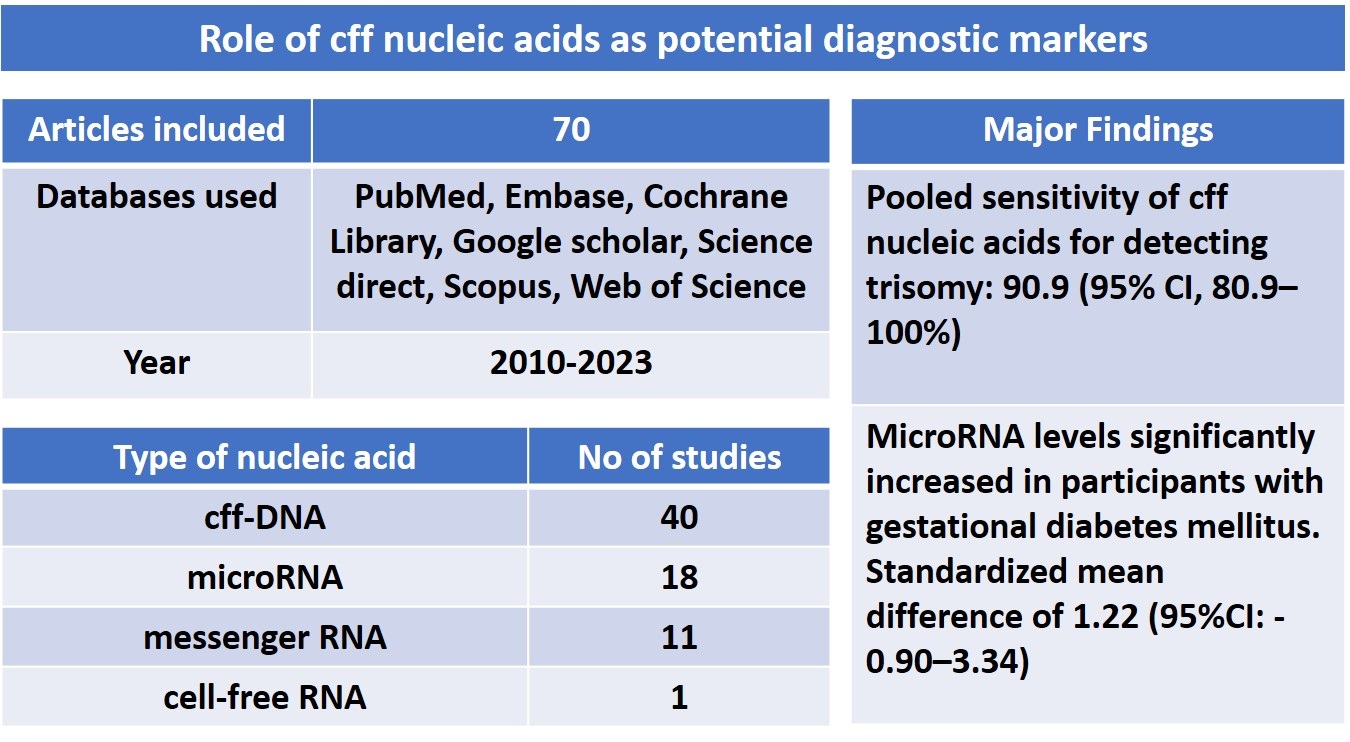
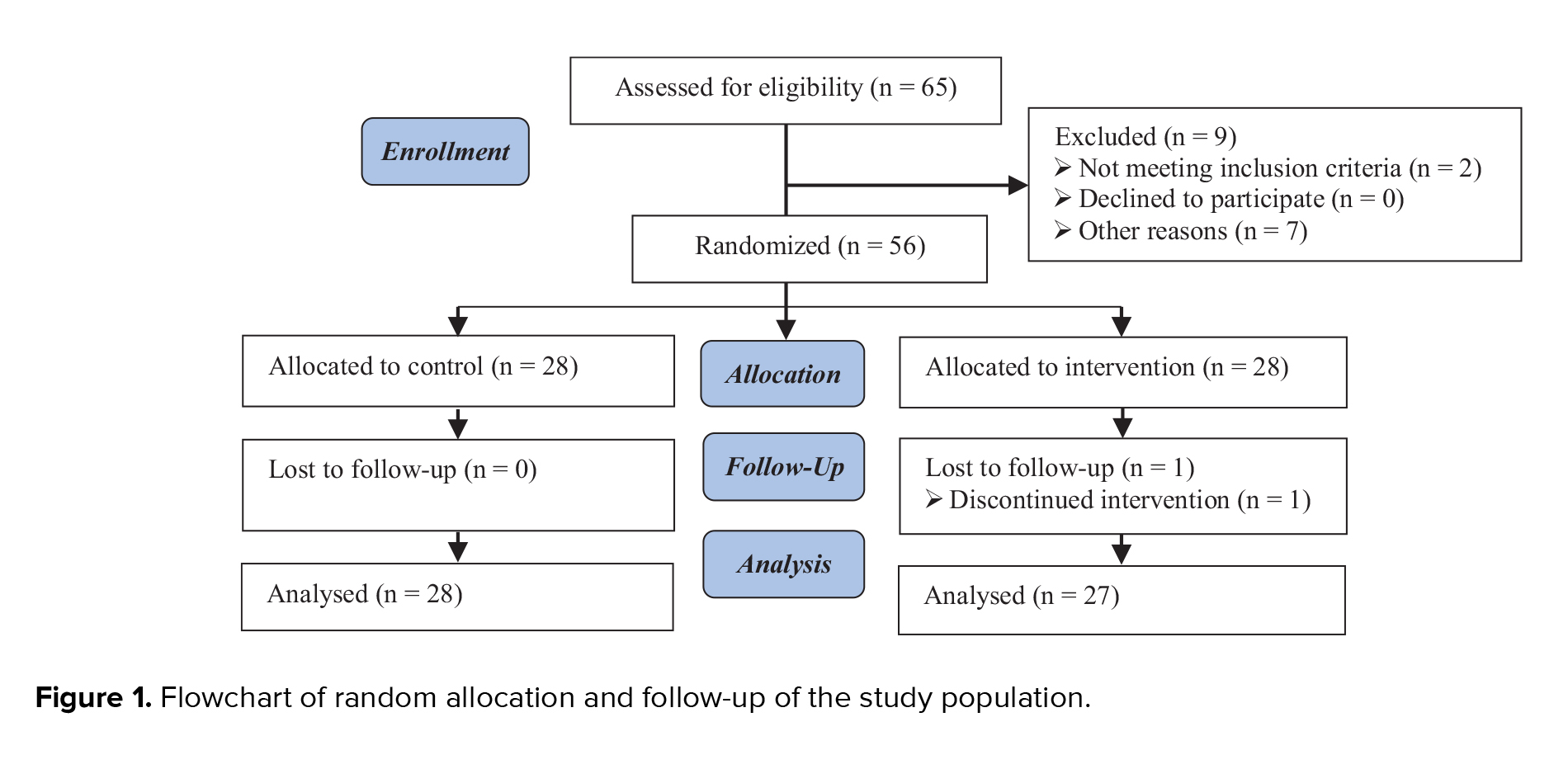
A total of 6099 citations were identified, with 2122 duplicate studies removed. These comprised 1104 from PubMed, 1239 from Embase, 145 from the Cochrane Library, 932 from Google Scholar, 766 from Scopus, 1088 from Science Direct, and 825 from Web of Science. Following the evaluation of titles and abstracts of 3977 articles, 2917 studies were excluded. Subsequently, 1060 articles underwent full-text review. After applying exclusion criteria, 990 complete texts were eliminated, leaving 70 articles for the final qualitative analysis. The study selection procedure is depicted in figure 1. This review, which includes 70 studies, aimed to determine whether the cff nucleic acid identifier markers in maternal circulation can be used as diagnostic risk factors for fetal outcomes. The publishing years were 2010-2023. Among 70 studies, 32 were prospective and retrospective cohorts, 29 were case-control, 08 were cross-sectional, and one study was randomized clinical trial. Most of the studies were conducted in the USA (n = 20), and among the remaining studies, 11 were from China, 7 from Italy, 4 from Australia, 3 each from the Czech Republic and Iran, 2 each from the UK, Denmark, Egypt, and Spain, 1 each from Belgium, Chile, Columbia, India, Israel, Japan, Malaysia, Mexico, Netherlands, Pakistan, Russia, South Africa, South Korea, and Turkey. The extracted data of the included studies are presented in table II.
3.2. Cff nucleic acids and pregnancy complications
Among 70 studies, 8 studies included subjects with fetal aneuploidy, 12 studies with FGR, 11 studies with gestational diabetes mellitus (GDM), 6 studies with pregnancy-induced hypertension, 19 studies with preterm birth (PTB), 24 studies with PE, 4 studies with spontaneous abortion, and 6 studies with subjects having morbidly adherent placenta. The cff nucleic acids included were fetal DNA (n = 40), miRNA (n = 18), mRNA (n = 11), and cfRNA (n = 1).
Using maternal blood cfDNA analysis, trisomies 21, 18, and 13 may be reliably screened for. In our study, which adopted a random-effects model, the pooled sensitivity was 90.9 (95% CI: 80.9-100). In the meta-analysis (Figure 2), it was determined that the weighted pooled detection rate (DR) for the total number of 1963 cases of trisomy 21, 563 cases of trisomy 18, and 119 cases of trisomy 13 was 99.7% (95% CI: 99.1-99.9), 98.2% (95% CI: 95.5-99.2), and 99.0% (95% CI: 65.8-100), respectively. This led to a false-positive rate of about 0.1% overall among the over 200,000 pregnancies that were not impacted by these trisomies. This study revealed significantly increased miRNA levels in participants with GDM; the standardized mean difference was 1.22 (95% CI: -0.90-3.34) (Figure 3). However, the variability was high (I2 = 97.8%). There have been fewer studies exploring the relationship between cffDNA and GDM, but the results have been mixed. Another association between low cffDNA percentage and an elevated risk of GDM has been found in the first and second trimesters.
24 studies investigated first and second-trimester levels of cff nucleic acids; most of these studies showed a statistically significant increase in cfDNA concentrations in women who went on to develop PE. Following the analysis of circulating miRNA expression patterns in the 4 participants with PE, it was postulated that miRNA might be useful in both the diagnosis and prognosis of PE. 43 miRNAs had a pooled sensitivity of 0.86 (95% CI: 0.81-0.90), specificity of 0.89 (95% CI: 0.85-0.92), and an area under the curve (AUC) of 0.94 (95% CI: 0.91-0.96) in 15 trials with 2042 healthy controls and 2685 PE cases. It was also observed that preterm delivery (PTD)-affected women had higher levels compared to controls who were matched for gestational age and delivered infants at term. It was found that in both high and low-risk pregnancies, cffDNA testing in the first or second trimester eliminated any possible predictive value for premature birth. The incidence of PTD in a group at high risk due to early contractions or a history of spontaneous PTD was identified using a panel of 7 mRNAs that were analyzed between 24 and 28 gestational weeks. In the fetuses of individuals who gave birth to premature twins, certain miRNA signatures have been longitudinally detected throughout gestation. Regrettably, there is no way to verify the results because the miRNAs found and looked at in each investigation have differed.
Yet, only a small number of research have examined the connection and possible positive predictive value (PPV) of cffDNA levels for FGR. It is possible that FGR women's elevated DNase activity, relative to unaffected women, significantly affects their levels of cffDNA. Overall cfDNA values were observed to be higher in FGR cases throughout the third trimester. Few studies have looked closely at the amount of cffDNA in morbidly adherent placentas. The morbidly connected placenta in 7 instances in the third trimester had no aberrant cfDNA.
3.3. Study quality assessment and publication bias
2 independent reviewers assessed the quality of each included study. Most studies exhibited a low to moderate risk of bias, with a high percentage of positive responses to the Joanna Briggs Institute's tool questions. However, 6 studies were identified with a high risk of bias (Figure 4). Visual inspection of the funnel plots suggested asymmetry. Nevertheless, Egger’s test indicated weak evidence for small-study effects (p > 0.05).










4. Discussion
Prenatal diagnostics and complicated pregnancies are 2 areas of obstetrical study that have promoted the use of cffDNA, a novel and promising biomarker. It is commonly known that circulating amounts of cffDNA contribute to several complications during pregnancy. While intriguing in vivo studies show the possibility of other processes, cffDNA is produced on trophoblastic cells as a result of apoptotic mechanisms. Because it occurs before the illness exhibits clinical signs, the rise in cffDNA levels is highly valuable for prenatal diagnosis as well as for predicting frequent complications associated with pregnancy. The available research was compiled in this systematic review to determine whether circulating nucleic acids in mother plasma and serum could function as additional, independent markers for the prediction and/or monitoring of the most common, severe pregnancy complications.
Reliable trisomies 21, 18, and 13 screening is provided by maternal blood cfDNA analysis. In the meta-analysis, it was determined that the weighted pooled DR for the total number of 1963 cases of trisomy 21, 563 cases of trisomy 18, and 119 cases of trisomy 13 was 99.7%, 98.2%, and 99.0%, respectively. The false-positive rate appeared to be equally low, although the reported DR of trisomy 21 by cfDNA testing was high but less than that in singleton pregnancies, as noted in an updated meta-analysis in twin pregnancies (82). Trisomy 21 was detected in 41 out of 42 pregnancies, reaching a 97.6% DR (17). The distinction between euploid and aneuploid pregnancy loss may be possible using cffDNA-based diagnostics, according to recent research (16).
During pregnancy, GDM is a temporary abnormal glucose tolerance that is identified in the latter half or third trimester of the pregnancy. Complications from GDM are quite likely to affect both the mother and the offspring. It has been proposed that circulating RNAs, particularly miRNAs, may be potential biomarkers for the early identification of GDM. MiRNA levels in participants with GDM were found to be considerably higher in this investigation; the standardized mean difference was 1.22 (95% CI: -0.90-3.34). According to a recent meta-analysis, circulating levels of miR-132 and miR-155 were lowered in participants with GDM, whereas miR-29a, miR-330, miR-134, miR-16, miR-223, and miR-17 showed substantial overall increase in GDM (83). With an odds ratio of 3.24, reduced expression of miR-20a-5p in females with one or more biophysical risk factors was found to be a strong predictor of GDM (40). Another study in 41 individuals during the first trimester discovered that pregnancies that would develop GDM had approximately 1.7-fold higher values for miR-125b-5p compared to the control group (43). A few studies have explored the relationship between cffDNA and GDM, but the findings have been inconsistent. While one study observed that cffDNA measured in the first trimester was significantly lower in women who later developed GDM (49), another found no difference in the second or third trimester (62). Additionally, low cffDNA percentages during the first and second trimesters have been linked to increased GDM risk in other studies (50, 26). Conversely, elevated levels were observed in women over 35 affected by GDM (23).
A pregnancy condition known as PE is characterized by an onset of proteinuria and hypertension after 20 wk of gestation. According to recent studies, genetic indicators could influence the likelihood of PE (67). In this study, a statistically significant rise in cfDNA concentrations were found in women who went on to develop PE. However, one study concluded that cfDNA is not a useful marker for PE after analyzing a large dataset of over 5000 cases (38). In contrast, after evaluating the circulating miRNA expression patterns in 4 participants with PE, it was hypothesized that miRNA could be beneficial for both the diagnosis and prognosis of PE (71). Another study found significant and consistent differences in cfRNA gene expression between normotensive and pre-eclamptic mothers early in gestation, well before symptoms appear (8). In 15 studies involving 2042 healthy controls and 2685 participants with PE, 43 miRNAs showed a pooled sensitivity of 0.86 (95% CI: 0.81-0.90), specificity of 0.89 (95% CI: 0.85-0.92), and an AUC of 0.94 (95% CI: 0.91-0.96). Another analysis from 8 papers, including 343 normal pregnancies and 273 participants with PE, revealed a diagnostic odds ratio of 50.24 (95% CI: 21.28-118.62), specificity of 0.87 (95% CI: 0.78-0.92), and pooled sensitivity of 0.88 (95% CI: 0.80-0.93). They concluded that there is a reasonably good diagnostic and predictive accuracy in using circulating miRNAs to differentiate participants with PE from controls (10).
An early delivery, occurring before 37 wk of gestation, is referred to as PTD. The majority of PTDs are spontaneous, resulting from preterm labor or preterm premature rupture of membranes. It has been proposed that maternal plasma cffDNA could serve as a marker for preterm labor. One study found that increased cffDNA levels at 14-20 wk were associated with a higher risk of PTD at both < 34 wk and < 37 wk (51); however, other studies did not identify any connection between cffDNA and PTD during the second trimester (32, 53). Similarly, another study found that PTD at < 37 wk was linked to cfDNA levels in the second trimester (50). Additionally, a recent study in China reported an inverse correlation between body mass index, maternal age, and the cell-free proportion of fetal DNA.
A panel of 7 mRNAs analyzed at 24-28 gestational weeks was found to detect the incidence of PTD in a high-risk group with early contractions or a history of spontaneous PTD (41). Certain miRNA signatures have been longitudinally identified during gestation in the fetuses of those who experienced a PTD birth. Unfortunately, the miRNAs identified and examined in each study have varied, and there is no way to validate the findings.
FGR is a diverse clinical syndrome linked to a higher risk of perinatal morbidity and death as well as fetal deterioration. The primary source of the increased cffDNA seen in participants with symptomatic FGR has been identified as placental malfunction, which is characteristic of FGR. Although limited studies have looked at the relationship and potential PPV of cffDNA levels for FGR. DNase activity, which is higher in FGR women than in unaffected women, may have a major impact on cffDNA levels in these women, according to a recent cohort study (47). The relationship between FGR and total DNA was examined in very few studies. In FGR instances, Muñoz-Hernández et al. (46) discovered greater total cfDNA readings throughout the third trimester. A single observation found that increased cfDNA was associated with FGR (24), but another study did not find a predictive effect of total cfDNA for FGR in the first trimester (65). Additionally, a recent study indicated that circulating miRNAs assessed were not accurate indicators for FGR prediction during the first trimester (31).
Morbidly adherent placentas are a broad range of placental defects caused by abnormal trophoblast invasion into the uterine wall's myometrium. Although a number of sonographic characteristics associated with invasive placentation have been identified, they lack sensitivity and specificity. As a result, additional markers may be helpful for monitoring the disease's course and facilitating early identification. The quantity of cffDNA in morbidly adherent placenta has not been thoroughly studied. In a study involving 7 cases in the third trimester, no abnormal cfDNA was found in the morbidly adherent placenta (64). However, other studies reported that human placental lactogen mRNA levels were 3 times higher (81, 30), and higher levels of miR-518b were observed in a case-control study (29). More research is needed to determine whether miRNA plays a role in predicting and monitoring morbidly adherent placentas.
5. Conclusion
Maternal care would be greatly influenced by a non-invasive, sensitive test that might detect pregnant women who are at high risk of developing complications early in the pregnancy. In addition to increasing clinical monitoring and perhaps lowering maternal-fetal morbidity as well as mortality, it would present a chance to implement preventive treatments for a subset of participants. Cff nucleic acids, if looked at in the first trimester, may prove to be a reliable noninvasive diagnostic tool for predicting serious complications during pregnancy. To validate this relationship, additional research is necessary.
Data Availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author Contributions
JR. Keshari and P. Prakash designed the study and conducted the research. JR. Keshari, P. Prakash, SR. Sinha, P. Prakash, K. Rani, T. Aziz, and Sh. Shilpa monitored, evaluated, and analyzed the study results. Further, JR. Keshari, P. Prakash, SR. Sinha, P. Prakash, K. Rani, T. Aziz, and Sh. Shilpa reviewed the article. All authors approved the final manuscript and take responsibility for the integrity of the data.
Acknowledgments
The authors express their sincere appreciation to Dr. Bindey Kumar, Director, IGIMS, Patna, Bihar, India, for his motivation to undertake this systematic review and meta-analysis. We also acknowledge Papyruscript Private Limited (https://www.papyruscript.com/) for their professional assistance in conducting the systematic review and meta-analysis, as well as for providing English language editing services.
Conflict of Interest
The authors declare that there is no conflict of interest.
In antenatal care, prenatal screening and diagnosis are routinely provided. It is believed that such amenities help women make decisions about whether to continue with pregnancies that are affected by developmental abnormalities. At present, the gold standard for prenatal diagnosis is the invasive method of acquiring fetal tissue and DNA. Though there is a little but considerable chance of miscarriage associated with invasive testing, many women prefer not to have it done (1, 2). To lower this risk of miscarriage and enable early testing, a reliable and feasible noninvasive prenatal diagnostic approach has long been sought upon.
The detection of cell-free fetal (cff) nucleic acids in the maternal bloodstream during pregnancy has completely changed the landscape of noninvasive prenatal screening and diagnosis. Circulating nucleic acids in maternal plasma and serum were anticipated to become a new area of interest in prenatal diagnostics following their discovery in 1997 (3). Although new in vivo studies show the possibility of other processes, cffDNA is released on trophoblastic cells as a result of apoptotic mechanisms. Since the rise in cffDNA levels occurs before the disease's clinical signs, it is highly valuable in the field of prenatal diagnosis and in predicting frequent pregnancy complications. Prenatal care has quickly included noninvasive prenatal screening utilizing cell-free DNA (cfDNA) in identifying common fetal aneuploidies, since its debut in 2011. According to a recent systematic review, the best sensitive and specific screening test for fetal Down syndrome, T13, and T18 in singleton and twin pregnancies is non-invasive prenatal screening with cfDNA. It was able to detect maternal diseases such as aneuploidies and malignancies, in contrast with conventional serum analyte screening (4).
While circulating cfDNA was the first known circulating nucleic acid, more have been discovered as molecular biology research and technology have advanced. At present, we take into consideration circulating nucleic acids in maternal plasma and serum, which is an array of extracellular nucleic acids that includes placental RNA, messenger RNA (mRNA), microRNA (miRNA), and other novel species like circular RNA, long noncoding RNA, mitochondrial DNA, and cfDNA (5). Alternative methods have been continuously explored, such as measuring total cfDNA or using gender-independent fetal epigenetic markers like DNA methylation and cfRNA of placental origin. These offer an appropriate replacement for genetic disease screening and prenatal diagnosis (6).
Recent research has demonstrated a correlation between elevated levels of cffDNA in maternal blood and various pregnancy complications, such as early pregnancy loss and preterm labor (7), pre-eclampsia (PE) (8-10), and fetal growth restriction (FGR) (11). Pregnancy difficulties have prompted studies to investigate the possibility of using cffDNA concentrations as a biomarker (12, 13). Research on the potential pathways connecting cffDNA to the etiology of pregnancy-related problems is only getting started (13).
We conducted a comprehensive review and meta-analysis to determine if the cff nucleic acid identification markers in the maternal bloodstream may be identified as diagnostic risk factors for fetal outcomes.
2. Materials and Methods
2.1. Search strategy
A comprehensive literature search was conducted in 7 different databases (PubMed, The Cochrane Library, Embase, Science Direct, Web of Science, Scopus, and Google Scholar). Preferred reporting items for systematic review and meta-analyses (PRISMA) checklist guidelines were followed (14). Articles published between 2010 and October 2023 were included in the study. During the literature search, restrictions were not placed on country, time, or language of publication. Editorial letters, conference proceedings, and practice guidelines were excluded.
The following key terms were used to identify relevant studies: cffDNA, next-generation sequencing (NGS), ctDNA, fetal outcomes, fetal anomalies, cffDNA, cff nucleic acids, epigenetic methylation, noninvasive prenatal diagnosis, prenatal diagnosis, and genetic diagnosis. Only research articles were retrieved and reviewed. All possible combinations of keywords were utilized in the search. Table I shows the search strings used to retrieve research articles from each database. After removing duplicates, titles and abstracts were screened as per eligibility criteria. Full-text articles of all the identified abstracts were reviewed independently.
2.2. Criteria for considering studies
Inclusion criteria encompass published research (case-control, cross-sectional, prospective, and retrospective cohorts) examining cff nucleic acid markers in maternal circulation. Studies on adult women ( ≥ 18 yr) with pregnancy-related complications were only included. Exclusion criteria encompass studies lacking relevance or adequate data, non-English publications, and certain study types (e.g., reviews, meta-analyses, letters to editors, commentaries, guidelines, book chapters, and editorials). Both published and unpublished studies, including gray literature, were considered for inclusion. The review also screened reference lists of prior systematic reviews/meta-analyses. Details such as authors, year of publication, country, study design, sample size, cell-free nucleic acid involved, pregnancy-related complications, and important results relevant to the study objectives were extracted from the included studies. The meta-analysis included studies based on specific criteria: relevant study design, availability of comparable outcome measures, sufficient statistical data, methodological quality, similar populations and interventions, and peer-reviewed full-text publications.
2.3. Study quality assessment
The Joanna Briggs Institute's critical appraisal checklists for studies were used to assess the quality of selected studies (15). A study was deemed to have a high risk of bias if the "yes" score was 49% or lower, moderate risk if the score was between 50 and 69%, and low risk if the score was 70% or higher. Each study underwent bias assessment and classification as low risk, high risk, or with some concerns. Any discrepancies between independent reviewers were resolved by consulting a third reviewer.
2.4. Statistical analysis and meta-analysis
For this meta-analysis, STATA version 17 was utilized. Continuous data were presented as means or medians with appropriate measures of variability. Categorical variables were expressed as numbers and percentages. A pooled meta-analysis was conducted. Heterogeneity was evaluated using Cochran’s Q test and the DerSimonian and Laird method. Heterogeneity levels were categorized as low, moderate, or high based on I-square values. Low heterogeneity was defined as an I-square value of < 25%, moderate heterogeneity as 25-50%, and high heterogeneity as > 50%. Publication bias was assessed visually with funnel plots and quantitatively using Egger’s test.


3. Results
3.1. Identification and description of studies
A total of 6099 citations were identified, with 2122 duplicate studies removed. These comprised 1104 from PubMed, 1239 from Embase, 145 from the Cochrane Library, 932 from Google Scholar, 766 from Scopus, 1088 from Science Direct, and 825 from Web of Science. Following the evaluation of titles and abstracts of 3977 articles, 2917 studies were excluded. Subsequently, 1060 articles underwent full-text review. After applying exclusion criteria, 990 complete texts were eliminated, leaving 70 articles for the final qualitative analysis. The study selection procedure is depicted in figure 1. This review, which includes 70 studies, aimed to determine whether the cff nucleic acid identifier markers in maternal circulation can be used as diagnostic risk factors for fetal outcomes. The publishing years were 2010-2023. Among 70 studies, 32 were prospective and retrospective cohorts, 29 were case-control, 08 were cross-sectional, and one study was randomized clinical trial. Most of the studies were conducted in the USA (n = 20), and among the remaining studies, 11 were from China, 7 from Italy, 4 from Australia, 3 each from the Czech Republic and Iran, 2 each from the UK, Denmark, Egypt, and Spain, 1 each from Belgium, Chile, Columbia, India, Israel, Japan, Malaysia, Mexico, Netherlands, Pakistan, Russia, South Africa, South Korea, and Turkey. The extracted data of the included studies are presented in table II.
3.2. Cff nucleic acids and pregnancy complications
Among 70 studies, 8 studies included subjects with fetal aneuploidy, 12 studies with FGR, 11 studies with gestational diabetes mellitus (GDM), 6 studies with pregnancy-induced hypertension, 19 studies with preterm birth (PTB), 24 studies with PE, 4 studies with spontaneous abortion, and 6 studies with subjects having morbidly adherent placenta. The cff nucleic acids included were fetal DNA (n = 40), miRNA (n = 18), mRNA (n = 11), and cfRNA (n = 1).
Using maternal blood cfDNA analysis, trisomies 21, 18, and 13 may be reliably screened for. In our study, which adopted a random-effects model, the pooled sensitivity was 90.9 (95% CI: 80.9-100). In the meta-analysis (Figure 2), it was determined that the weighted pooled detection rate (DR) for the total number of 1963 cases of trisomy 21, 563 cases of trisomy 18, and 119 cases of trisomy 13 was 99.7% (95% CI: 99.1-99.9), 98.2% (95% CI: 95.5-99.2), and 99.0% (95% CI: 65.8-100), respectively. This led to a false-positive rate of about 0.1% overall among the over 200,000 pregnancies that were not impacted by these trisomies. This study revealed significantly increased miRNA levels in participants with GDM; the standardized mean difference was 1.22 (95% CI: -0.90-3.34) (Figure 3). However, the variability was high (I2 = 97.8%). There have been fewer studies exploring the relationship between cffDNA and GDM, but the results have been mixed. Another association between low cffDNA percentage and an elevated risk of GDM has been found in the first and second trimesters.
24 studies investigated first and second-trimester levels of cff nucleic acids; most of these studies showed a statistically significant increase in cfDNA concentrations in women who went on to develop PE. Following the analysis of circulating miRNA expression patterns in the 4 participants with PE, it was postulated that miRNA might be useful in both the diagnosis and prognosis of PE. 43 miRNAs had a pooled sensitivity of 0.86 (95% CI: 0.81-0.90), specificity of 0.89 (95% CI: 0.85-0.92), and an area under the curve (AUC) of 0.94 (95% CI: 0.91-0.96) in 15 trials with 2042 healthy controls and 2685 PE cases. It was also observed that preterm delivery (PTD)-affected women had higher levels compared to controls who were matched for gestational age and delivered infants at term. It was found that in both high and low-risk pregnancies, cffDNA testing in the first or second trimester eliminated any possible predictive value for premature birth. The incidence of PTD in a group at high risk due to early contractions or a history of spontaneous PTD was identified using a panel of 7 mRNAs that were analyzed between 24 and 28 gestational weeks. In the fetuses of individuals who gave birth to premature twins, certain miRNA signatures have been longitudinally detected throughout gestation. Regrettably, there is no way to verify the results because the miRNAs found and looked at in each investigation have differed.
Yet, only a small number of research have examined the connection and possible positive predictive value (PPV) of cffDNA levels for FGR. It is possible that FGR women's elevated DNase activity, relative to unaffected women, significantly affects their levels of cffDNA. Overall cfDNA values were observed to be higher in FGR cases throughout the third trimester. Few studies have looked closely at the amount of cffDNA in morbidly adherent placentas. The morbidly connected placenta in 7 instances in the third trimester had no aberrant cfDNA.
3.3. Study quality assessment and publication bias
2 independent reviewers assessed the quality of each included study. Most studies exhibited a low to moderate risk of bias, with a high percentage of positive responses to the Joanna Briggs Institute's tool questions. However, 6 studies were identified with a high risk of bias (Figure 4). Visual inspection of the funnel plots suggested asymmetry. Nevertheless, Egger’s test indicated weak evidence for small-study effects (p > 0.05).











4. Discussion
Prenatal diagnostics and complicated pregnancies are 2 areas of obstetrical study that have promoted the use of cffDNA, a novel and promising biomarker. It is commonly known that circulating amounts of cffDNA contribute to several complications during pregnancy. While intriguing in vivo studies show the possibility of other processes, cffDNA is produced on trophoblastic cells as a result of apoptotic mechanisms. Because it occurs before the illness exhibits clinical signs, the rise in cffDNA levels is highly valuable for prenatal diagnosis as well as for predicting frequent complications associated with pregnancy. The available research was compiled in this systematic review to determine whether circulating nucleic acids in mother plasma and serum could function as additional, independent markers for the prediction and/or monitoring of the most common, severe pregnancy complications.
Reliable trisomies 21, 18, and 13 screening is provided by maternal blood cfDNA analysis. In the meta-analysis, it was determined that the weighted pooled DR for the total number of 1963 cases of trisomy 21, 563 cases of trisomy 18, and 119 cases of trisomy 13 was 99.7%, 98.2%, and 99.0%, respectively. The false-positive rate appeared to be equally low, although the reported DR of trisomy 21 by cfDNA testing was high but less than that in singleton pregnancies, as noted in an updated meta-analysis in twin pregnancies (82). Trisomy 21 was detected in 41 out of 42 pregnancies, reaching a 97.6% DR (17). The distinction between euploid and aneuploid pregnancy loss may be possible using cffDNA-based diagnostics, according to recent research (16).
During pregnancy, GDM is a temporary abnormal glucose tolerance that is identified in the latter half or third trimester of the pregnancy. Complications from GDM are quite likely to affect both the mother and the offspring. It has been proposed that circulating RNAs, particularly miRNAs, may be potential biomarkers for the early identification of GDM. MiRNA levels in participants with GDM were found to be considerably higher in this investigation; the standardized mean difference was 1.22 (95% CI: -0.90-3.34). According to a recent meta-analysis, circulating levels of miR-132 and miR-155 were lowered in participants with GDM, whereas miR-29a, miR-330, miR-134, miR-16, miR-223, and miR-17 showed substantial overall increase in GDM (83). With an odds ratio of 3.24, reduced expression of miR-20a-5p in females with one or more biophysical risk factors was found to be a strong predictor of GDM (40). Another study in 41 individuals during the first trimester discovered that pregnancies that would develop GDM had approximately 1.7-fold higher values for miR-125b-5p compared to the control group (43). A few studies have explored the relationship between cffDNA and GDM, but the findings have been inconsistent. While one study observed that cffDNA measured in the first trimester was significantly lower in women who later developed GDM (49), another found no difference in the second or third trimester (62). Additionally, low cffDNA percentages during the first and second trimesters have been linked to increased GDM risk in other studies (50, 26). Conversely, elevated levels were observed in women over 35 affected by GDM (23).
A pregnancy condition known as PE is characterized by an onset of proteinuria and hypertension after 20 wk of gestation. According to recent studies, genetic indicators could influence the likelihood of PE (67). In this study, a statistically significant rise in cfDNA concentrations were found in women who went on to develop PE. However, one study concluded that cfDNA is not a useful marker for PE after analyzing a large dataset of over 5000 cases (38). In contrast, after evaluating the circulating miRNA expression patterns in 4 participants with PE, it was hypothesized that miRNA could be beneficial for both the diagnosis and prognosis of PE (71). Another study found significant and consistent differences in cfRNA gene expression between normotensive and pre-eclamptic mothers early in gestation, well before symptoms appear (8). In 15 studies involving 2042 healthy controls and 2685 participants with PE, 43 miRNAs showed a pooled sensitivity of 0.86 (95% CI: 0.81-0.90), specificity of 0.89 (95% CI: 0.85-0.92), and an AUC of 0.94 (95% CI: 0.91-0.96). Another analysis from 8 papers, including 343 normal pregnancies and 273 participants with PE, revealed a diagnostic odds ratio of 50.24 (95% CI: 21.28-118.62), specificity of 0.87 (95% CI: 0.78-0.92), and pooled sensitivity of 0.88 (95% CI: 0.80-0.93). They concluded that there is a reasonably good diagnostic and predictive accuracy in using circulating miRNAs to differentiate participants with PE from controls (10).
An early delivery, occurring before 37 wk of gestation, is referred to as PTD. The majority of PTDs are spontaneous, resulting from preterm labor or preterm premature rupture of membranes. It has been proposed that maternal plasma cffDNA could serve as a marker for preterm labor. One study found that increased cffDNA levels at 14-20 wk were associated with a higher risk of PTD at both < 34 wk and < 37 wk (51); however, other studies did not identify any connection between cffDNA and PTD during the second trimester (32, 53). Similarly, another study found that PTD at < 37 wk was linked to cfDNA levels in the second trimester (50). Additionally, a recent study in China reported an inverse correlation between body mass index, maternal age, and the cell-free proportion of fetal DNA.
A panel of 7 mRNAs analyzed at 24-28 gestational weeks was found to detect the incidence of PTD in a high-risk group with early contractions or a history of spontaneous PTD (41). Certain miRNA signatures have been longitudinally identified during gestation in the fetuses of those who experienced a PTD birth. Unfortunately, the miRNAs identified and examined in each study have varied, and there is no way to validate the findings.
FGR is a diverse clinical syndrome linked to a higher risk of perinatal morbidity and death as well as fetal deterioration. The primary source of the increased cffDNA seen in participants with symptomatic FGR has been identified as placental malfunction, which is characteristic of FGR. Although limited studies have looked at the relationship and potential PPV of cffDNA levels for FGR. DNase activity, which is higher in FGR women than in unaffected women, may have a major impact on cffDNA levels in these women, according to a recent cohort study (47). The relationship between FGR and total DNA was examined in very few studies. In FGR instances, Muñoz-Hernández et al. (46) discovered greater total cfDNA readings throughout the third trimester. A single observation found that increased cfDNA was associated with FGR (24), but another study did not find a predictive effect of total cfDNA for FGR in the first trimester (65). Additionally, a recent study indicated that circulating miRNAs assessed were not accurate indicators for FGR prediction during the first trimester (31).
Morbidly adherent placentas are a broad range of placental defects caused by abnormal trophoblast invasion into the uterine wall's myometrium. Although a number of sonographic characteristics associated with invasive placentation have been identified, they lack sensitivity and specificity. As a result, additional markers may be helpful for monitoring the disease's course and facilitating early identification. The quantity of cffDNA in morbidly adherent placenta has not been thoroughly studied. In a study involving 7 cases in the third trimester, no abnormal cfDNA was found in the morbidly adherent placenta (64). However, other studies reported that human placental lactogen mRNA levels were 3 times higher (81, 30), and higher levels of miR-518b were observed in a case-control study (29). More research is needed to determine whether miRNA plays a role in predicting and monitoring morbidly adherent placentas.
5. Conclusion
Maternal care would be greatly influenced by a non-invasive, sensitive test that might detect pregnant women who are at high risk of developing complications early in the pregnancy. In addition to increasing clinical monitoring and perhaps lowering maternal-fetal morbidity as well as mortality, it would present a chance to implement preventive treatments for a subset of participants. Cff nucleic acids, if looked at in the first trimester, may prove to be a reliable noninvasive diagnostic tool for predicting serious complications during pregnancy. To validate this relationship, additional research is necessary.
Data Availability
Data supporting the findings of this study are available upon reasonable request from the corresponding author.
Author Contributions
JR. Keshari and P. Prakash designed the study and conducted the research. JR. Keshari, P. Prakash, SR. Sinha, P. Prakash, K. Rani, T. Aziz, and Sh. Shilpa monitored, evaluated, and analyzed the study results. Further, JR. Keshari, P. Prakash, SR. Sinha, P. Prakash, K. Rani, T. Aziz, and Sh. Shilpa reviewed the article. All authors approved the final manuscript and take responsibility for the integrity of the data.
Acknowledgments
The authors express their sincere appreciation to Dr. Bindey Kumar, Director, IGIMS, Patna, Bihar, India, for his motivation to undertake this systematic review and meta-analysis. We also acknowledge Papyruscript Private Limited (https://www.papyruscript.com/) for their professional assistance in conducting the systematic review and meta-analysis, as well as for providing English language editing services.
Conflict of Interest
The authors declare that there is no conflict of interest.
Type of Study: Review Article |
Subject:
Reproductive Genetics
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |