Fri, Jan 30, 2026
[Archive]
Volume 23, Issue 6 (June 2025)
IJRM 2025, 23(6): 517-526 |
Back to browse issues page
Ethics code: IR.IAU.VARAMIN.REC.1401.006
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Ashrafzadeh H, Tafvizi F, Ghasemi N, Vahidi Mehrjardi M Y, Naseh V. Prenatal diagnosis using next-generation sequencing in genetic counseling: Novel mutations in three large Iranian families: A case series. IJRM 2025; 23 (6) :517-526
URL: http://ijrm.ir/article-1-3576-en.html
URL: http://ijrm.ir/article-1-3576-en.html
Hamidreza Ashrafzadeh1 

 , Farzaneh Tafvizi1
, Farzaneh Tafvizi1 

 , Nasrin Ghasemi2
, Nasrin Ghasemi2 

 , Mohammad Yahya Vahidi Mehrjardi3
, Mohammad Yahya Vahidi Mehrjardi3 

 , Vahid Naseh *4
, Vahid Naseh *4 




 , Farzaneh Tafvizi1
, Farzaneh Tafvizi1 

 , Nasrin Ghasemi2
, Nasrin Ghasemi2 

 , Mohammad Yahya Vahidi Mehrjardi3
, Mohammad Yahya Vahidi Mehrjardi3 

 , Vahid Naseh *4
, Vahid Naseh *4 


1- Department of Biology, Pa.c., Islamic Azad University, Parand, Iran.
2- Abortion Research Center, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
3- Diabetes Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
4- Department of Biology, Pa.c., Islamic Azad University, Parand, Iran. ,vahid55vet@yahoo.com
2- Abortion Research Center, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
3- Diabetes Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
4- Department of Biology, Pa.c., Islamic Azad University, Parand, Iran. ,
Abstract: (422 Views)
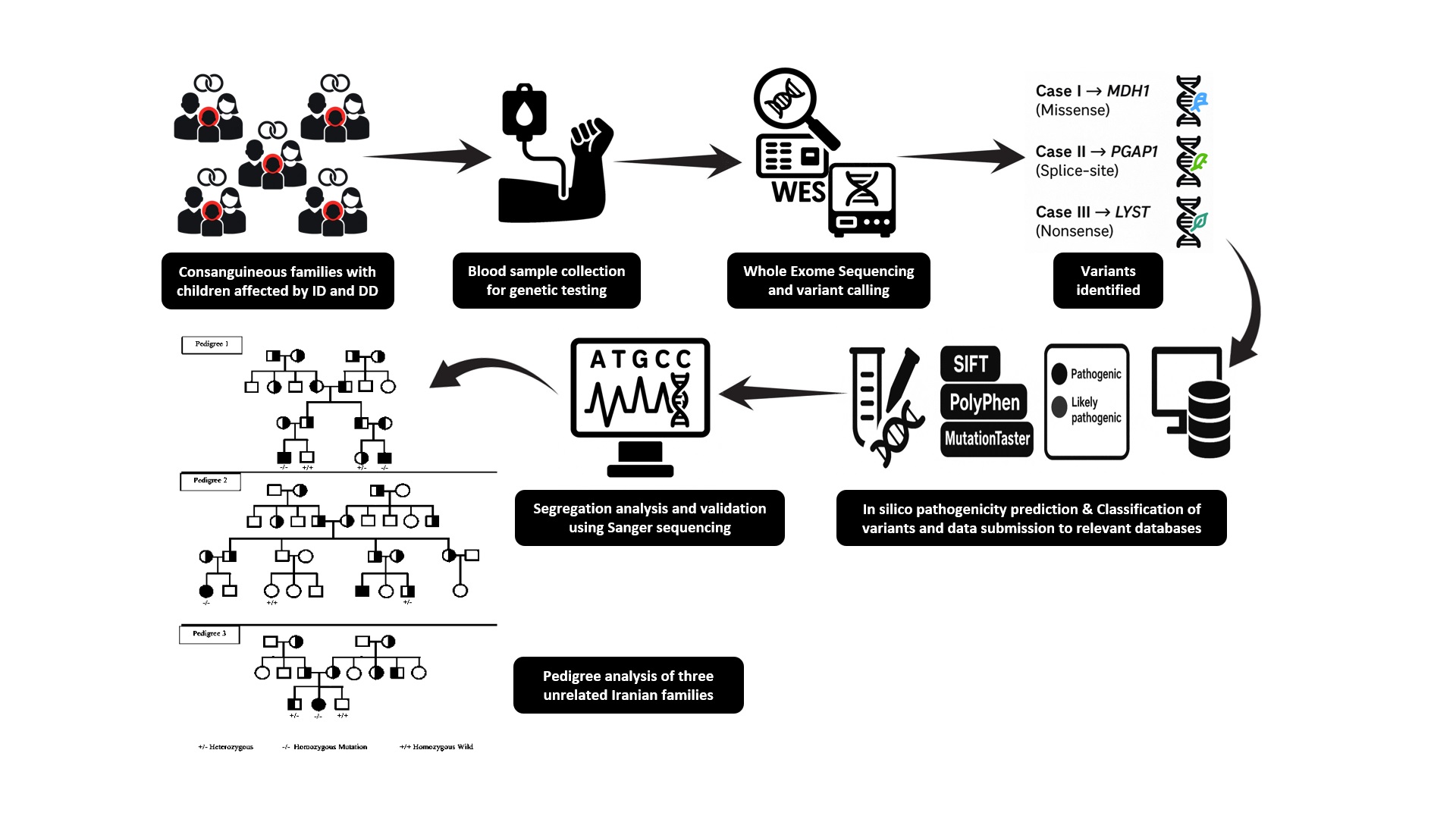
Background: All considerable families are seeking genetic counseling aiming to manage the next pregnancy according to the positive family history of heterogenetic disorders. Prenatal diagnosis utilizing next-generation sequencing provides a significant means to identify the causes of genetic abnormalities, allowing for timely interventions that support informed family planning. This study explores the power of whole-exome sequencing (WES) in uncovering genetic variants in couples who are seeking genetic counseling for their next pregnancy.
Case Presentations: In this study, WES was used to identify genetic variations associated with disability in families seeking genetic counseling. 3 families who had at least 1 child with developmental delay (DD) and/or intellectual disability (ID) participated in a genetic counselling clinic, Yazd Reproductive Science Institute, Yazd, Iran to have successful outcomes for the next pregnancy. 3 distinct mutation sites from 3 families were diagnosed, following the WES for affected children with intellectual disabilities. Results showed a homozygous de novo stop-gain mutation in malate dehydrogenase 1 gene (NM_005917.4:c.4C>T; p.Arg2Ter), a splice acceptor mutation in the post-glycosylphosphatidylinositol attachment to proteins inositol deacylase 1 gene (NM_024989.4:c.1221-1G>T), and a missense mutation in the lysosomal trafficking regulator gene (NM_000081.4:c.949G>A; p.Glu317Lys) in each family, respectively.
Conclusion: For cases with DD and unexplained ID, WES is a very successful diagnostic approach. Unfortunately, large Iranian families exhibit significant genetic heterogeneity, highlighting the critical role of de novo variants in diagnosis. The results of this study confirm that proteins inositol deacylase 1, malate dehydrogenase 1, and lysosomal trafficking regulator are involved in the pathophysiology of ID/DD and the transformative potential of prenatal genetic screening.
Case Presentations: In this study, WES was used to identify genetic variations associated with disability in families seeking genetic counseling. 3 families who had at least 1 child with developmental delay (DD) and/or intellectual disability (ID) participated in a genetic counselling clinic, Yazd Reproductive Science Institute, Yazd, Iran to have successful outcomes for the next pregnancy. 3 distinct mutation sites from 3 families were diagnosed, following the WES for affected children with intellectual disabilities. Results showed a homozygous de novo stop-gain mutation in malate dehydrogenase 1 gene (NM_005917.4:c.4C>T; p.Arg2Ter), a splice acceptor mutation in the post-glycosylphosphatidylinositol attachment to proteins inositol deacylase 1 gene (NM_024989.4:c.1221-1G>T), and a missense mutation in the lysosomal trafficking regulator gene (NM_000081.4:c.949G>A; p.Glu317Lys) in each family, respectively.
Conclusion: For cases with DD and unexplained ID, WES is a very successful diagnostic approach. Unfortunately, large Iranian families exhibit significant genetic heterogeneity, highlighting the critical role of de novo variants in diagnosis. The results of this study confirm that proteins inositol deacylase 1, malate dehydrogenase 1, and lysosomal trafficking regulator are involved in the pathophysiology of ID/DD and the transformative potential of prenatal genetic screening.
Keywords: Prenatal diagnosis, Genetic counselling, Whole exome sequencing, Intellectual disability, Developmental delay.
Type of Study: Case Report |
Subject:
Reproductive Genetics
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |